-
PDF
- Split View
-
Views
-
Cite
Cite
Lisa Prince, Malgorzata Korbas, Philip Davidson, Karin Broberg, Matthew Dearborn Rand, Target Organ Specific Activity of Drosophila MRP (ABCC1) Moderates Developmental Toxicity of Methylmercury, Toxicological Sciences, Volume 140, Issue 2, August 2014, Pages 425–435, https://doi.org/10.1093/toxsci/kfu095
Close - Share Icon Share
Abstract
Methylmercury (MeHg) is a ubiquitous and persistent neurotoxin that poses a risk to human health. Although the mechanisms of MeHg toxicity are not fully understood, factors that contribute to susceptibility are even less well known. Studies of human gene polymorphisms have identified a potential role for the multidrug resistance-like protein (MRP/ABCC) family, ATP-dependent transporters, in MeHg susceptibility. MRP transporters have been shown to be important for MeHg excretion in adult mouse models, but their role in moderating MeHg toxicity during development has not been explored. We therefore investigated effects of manipulating expression levels of MRP using a Drosophila development assay. Drosophila MRP (dMRP) is homologous to human MRP1–4 (ABCC1–4), sharing 50% identity and 67% similarity with MRP1. A greater susceptibility to MeHg is seen in dMRP mutant flies, demonstrated by reduced rates of eclosion on MeHg-containing food. Furthermore, targeted knockdown of dMRP expression using GAL4>UAS RNAi methods demonstrates a tissue-specific function for dMRP in gut, Malpighian tubules, and the nervous system in moderating developmental susceptibility to MeHg. Using X-ray synchrotron fluorescence imaging, these same tissues were also identified as the highest Hg-accumulating tissues in fly larvae. Moreover, higher levels of Hg are seen in dMRP mutant larvae compared with a control strain fed an equivalent dose of MeHg. In sum, these data demonstrate that dMRP expression, both globally and within Hg-targeted organs, has a profound effect on susceptibility to MeHg in developing flies. Our findings point to a potentially novel and specific role for dMRP in neurons in the protection against MeHg. Finally, this experimental system provides a tractable model to evaluate human polymorphic variants of MRP and other gene variants relevant to genetic studies of mercury-exposed populations.
Methylmercury (MeHg) is a ubiquitous and persistent developmental neurotoxin that occurs naturally in the environment and has been enhanced by anthropogenic sources. The mechanisms that make MeHg especially toxic during development remain uncertain, despite numerous laboratory studies using rodents, zebrafish, worms, and cell culture models. MeHg exposure through the consumption of naturally or anthropogenically contaminated fish and seafood is common in many human populations. The developing nervous system is especially sensitive to the toxic effects of MeHg. Large-scale epidemiological studies of children exposed prenatally to MeHg from maternal consumption of fish or other seafood have yielded inconsistent results regarding the presence or absence of abnormal developmental outcomes (Davidson et al., 1998; Grandjean et al., 1997). This inconsistency suggests that other factors such as genetics may contribute to individual or population differences in MeHg susceptibility (Julvez and Grandjean, 2013; NRC, 2000). Recently investigators have been exploring the genetic factors relating to MeHg toxicity by probing polymorphisms in genes presumed to be involved in its metabolism and excretion (Custodio et al., 2004; Engstrom et al., 2011, 2013). It is therefore important that the fundamental mechanisms of MeHg developmental toxicity are elaborated using tractable models to experimentally assess candidate genes involved in MeHg toxicity.
MeHg and its inorganic counterpart are commonly excreted as a conjugate with glutathione (GSH) (Cernichiari et al., 2007) implicating a role for conventional xenobiotic metabolism genes and pathways in its elimination. Consistent with this notion, several studies have demonstrated that efflux of toxic metals is reliant on multidrug resistance-like protein (MRP) transporters, which are members of the ATP-binding Cassette (ABC) superfamily that carry out Phase III xenobiotic metabolism, preferentially transporting substrates that are conjugated to GSH (Cole, 2014). Although best characterized in the cellular efflux of drugs, members of the MRP family have proven to facilitate efflux of mercury (Aleo et al., 2005; Bridges et al., 2013; Wortelboer et al., 2008). For example, pharmacological inhibition of MRP1 and MRP2 in kidney cells cultured with mercury results in elevated accumulation of mercury (Aleo et al., 2005). Relative to control mice, a strain carrying a null mutation in MRP2 (ABCC2) retains higher levels of inorganic mercury from an oral dose (Bridges et al., 2013). Furthermore, these same MRP2 null mice accumulate mercury to higher levels in multiple organs, including the placenta and fetus (Bridges et al., 2012). Importantly, human polymorphisms in MRP2 (ABCC2) are reported to correlate with differences in urinary excretion of inorganic mercury in an environmentally exposed population (Engstrom et al., 2013). Despite evidence that MRPs can influence MeHg elimination, the potential role of MRPs in moderating MeHg toxicity during critical stages of development has not been investigated.
In this study, we examine the role of Drosophila MRP (dMRP) in conferring resistance to MeHg toxicity during development. MeHg exposure to flies during the larval feeding stage shows a dose-responsive sensitivity of development through pupal metamorphosis to the adult stage (Mahapatra et al., 2010). In previous studies, we have demonstrated the efficacy of this model in identifying genes that functionally influence MeHg toxicity during development (Mahapatra et al., 2010; Rand et al., 2012). dMRP is highly homologous to human MRP1–4 (encoded by genes ABCC1–4) and shares the highest identity (50%) and similarity (67%) with human MRP1 (Fig. 3A, Grailles et al., 2003). dMRP has also been shown to be functionally similar to the human MRPs, including MRP1 (Szeri et al., 2009). We therefore investigated effects of manipulating endogenous expression levels of dMRP on the overall developmental outcome of larval flies exposed to MeHg. Our approach involved transgenic RNAi knockdown of dMRP in a tissue-specific manner to identify target organs that contribute to overall MeHg susceptibility. Moreover, we characterized the localization of Hg, from a MeHg treatment, in larval tissues to begin to correlate the functional effects of altering the expression of the MRP transporter in relevant organs targeted by MeHg.
Elemental localization with X-ray fluorescence in Drosophila larvae. (A, B) Larval anatomy in a paraffin section and in a schematic representation (B, adapted from Hartenstein (1993)). Br—brain; VNC—ventral nerve cord; SG—salivary gland; PV—proventriculus; GC—gastric caeca; MT—Malpighian tubule; MG—midgut; HG—hindgut; Cu—cuticle; FB—fat body (not illustrated in B). (C–F) X-ray fluorescence maps for mercury (Hg, C), iron (Fe, D), zinc (Zn, E), and calcium (Ca, F). Levels of detection are adjusted to minimum and maximum signal intensity across the sample as denoted by the heat map scale in each panel.
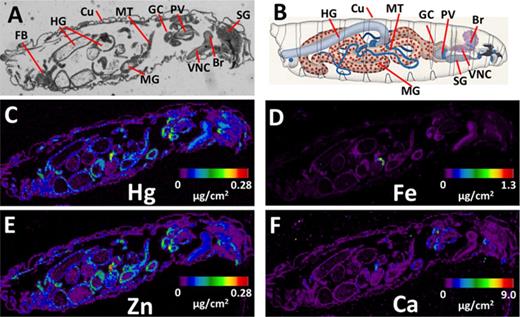
Distribution of Hg versus other elements in larval central nervous system (CNS). (A) X-ray fluorescence of Hg in the anterior region of the larva highlighting the CNS. (B) Immunostaining for the Elav neural specific antigen (red) shows labeling of the cellular cortical regions of the brain (Br) and ventral nerve cord (VNC). (C–D) Distribution of iron (Fe) and calcium (Ca) in the neuropil and cortex.
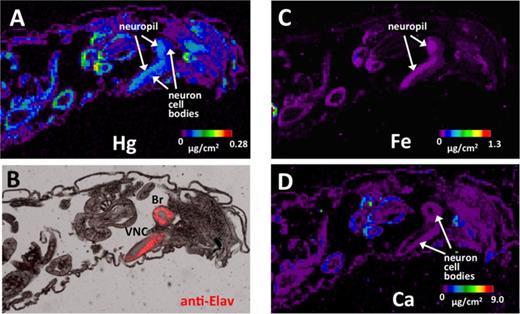
Characterization of dMRP mutant flies. (A) dMRP identity and similarity scores with human MRP paralogs (performed with NCBI Blast). (B) A schematic of the inserts in the MRPEY11919 and MRPMI00447 mutants. (C) Tolerance to MeHg was determined by eclosion assay with the two dMRP mutant strains, MRPEY11919 and MRPMI00447 in comparison to a control strain (y1w67c23, (yw)). Asterisks indicate significant difference between mutant strain and control strain at each treatment, as determined by a z-test (**p<0.001; ***p<0.0001). (D) qRT-PCR determination of dMRP transcript levels in RNA isolated from the two mutant strains, normalized to GAPDH1 and expressed relative to levels in the control strain (yw). Asterisks indicate significant difference between mutant line and control as determined by t-test (***p<0.0001). (E, G) Localization of dMRP expression by immunostaining of sections of third instar wandering larvae of the MRPMI00447 mutant strain that carries GFP expressed by the MRP enhancer. (F) Control immunostaining of a section of the MRPEY11919 that lacks the GFP reporter. (G) Close-up of fluorescence in midgut (MG), Malpighian tubules (MT), body wall muscle (MS), and fat body (FB) in MRPMI00447. (H) Differential Interference Contrast (DIC) image of panel (G).
MATERIALS AND METHODS
Fly strains
The following fly strains were obtained from the Bloomington Drosophila Stock Center at Indiana University (Bloomington, Indiana): Canton S (no. 1), Hikone R (no. 4267), FB-Gal4 (no. 6984), C42-Gal4 (no. 30835), Elav-Gal4 (no. 458), Repo-Gal4 (no. 7415), MRPEY11919 (no. 20712), y1w67c23 (no. 6599), and MRPMI00447 (no. 31007). The MRPEY11919 mutant carries a P-element insert at 2L:12722365 with an intron-less yellow+ gene and the mini-white gene in addition to a 3’ outwardly directed promoter and upstream activation sequence (UAS) (Bellen et al., 2004). The MRPMI00447 mutant carries a Minos-mediated integration cassette (MiMIC) that encodes a splice acceptor followed by a stop codon, which is followed by the green fluorescent protein (GFP) coding sequence and a yellow+ gene. This construct is flanked by ΦC31 attP integrase sites for downstream applications described in Venken et al. (2011). This insert lies at 2L:12721222 that disrupts eight of the 17 predicted splice variant transcripts of the MRP gene. The Repo-GAL4 insertion is homozygous lethal and was therefore maintained with a Serrate and Actin GFP marked balancer line for identification of flies that contained the Repo-GAL4 driver (GFP negative larvae) in crosses described below. Fly lines obtained from the Vienna Drosophila RNAi Center (Vienna, Austria) include UAS-dMRP RNAi (no. 105419) (Dietzl et al., 2007) and the yw1118 control strain (no. 60100). The NP1-GAL4 line was provided by Benoit Biteau (University of Rochester) and can be obtained from the Drosophila Genetic Resource at the National Institute of Genetics (Shizuoka, Japan).
All flies were kept at 25°C on standard fly food made of cornmeal, molasses, yeast, and agar. All crosses were performed between respective virgin female GAL4 lines and male UAS dMRP RNAi or its relevant control line (yw1118).
MeHg tolerance assays
Tolerance of the transgenic crosses to MeHg was determined by performing a previously described eclosion assay (Mahapatra et al., 2010). Briefly, first instar larvae were placed on vials of fly food containing 0, 5, 10, 15, or 20μM MeHg (Methylmercury chloride, Aldrich no. 442534). A total of 150 larvae were assayed, in three batches of 50 larvae each, for each MeHg concentration. Eclosion (emergence of adult flies from the pupal case) was scored on day 13 after larvae were placed on food. Higher eclosion percentages are indicative of greater tolerance to MeHg toxicity in this assay.
Statistical analyses
Rates of eclosion for indicated strains at each MeHg concentration are expressed in proportions (% eclosion). Assays were performed to achieve n = 150. Statistical consideration of differences between experimental and control fly strains are therefore comparisons of proportion values and not of continuous values. Because error determinations in proportion values become restricted at the edges (i.e., near 0% and 100%), an analysis of variance was not used. In addition, the highly threshold nature of the dose response was not suitable for standard probit analysis. Statistical analyses of eclosion assays were therefore done using a pairwise two-tailed z-test, treating each MeHg concentration categorically. p-values of less than 0.01 were considered significant. For dMRP gene expression in the MRPMI00447 and MRPEY11919 larvae using quantitative real-time PCR (qRT-PCR), where values are continuous and assays were performed in replicates of five, a pairwise t-test was used with p<0.05 considered significant.
Larval MeHg treatments, fixation, sectioning, and immunostaining
Second instar wild-type larvae (Canton S and Hikone R strains) were fed on food containing 15μM MeHg (or DMSO solvent control food) for 72 h at 25°C. Larvae were transferred to normal food for a period of 4 h to clear the MeHg containing food from the gut and external cuticle. Larvae were then rinsed in PBS and fixed by immersion in Carnoy's fixative (6:3:1 Ethanol:Chloroform:Acetic acid) for 1 h at room temperature. Larvae were directly transferred to 30% ethanol and dehydrated in 30%, 50%, and 70% ethanol incubations for 10 min each, and they were stored in 70% ethanol at 4°C until further processing. Analysis of Hg at this stage showed that at least 80% of Hg was retained in the larvae relative to larvae collected immediately prior to fixation (data not shown).
Embedding and sectioning was carried out in the University of Rochester Environmental Health Science Center Histology Core. Briefly, an ethanol (100%) wash was followed by xylene treatment and then paraffin embedding. Serial sectioning was done on a microtome set at 10 μm thickness and mounted on X-ray synchrotron fluorescence imaging (XFI)-compatible Thermanox plastic coverslips (EMA/Nunc no. 72285) or on Superfrost plus glass slides (Fisher no. 12–550–15).
Immunostaining with the pan-neural antibody anti-Elav (1:10, 9F8A9, Developmental Studies Hybridoma Bank, University of Iowa) was performed on xylene cleared sections on Thermanox coverslips. An adjacent section to that used for XFI was used for immunostaining. Alexa555-conjugated goat anti-mouse secondary antibody (1:400, Jackson Immunoresearch) was used and sections were imaged on a Nikon AZ100 microscope. Immunolocalization of dMRP expression was done with sections of MRPMI00447 and control (MRPEY11919) larvae with no MeHg treatment. GFP reporter expression was revealed with rabbit anti-GFP (1:500, no. TP401, Torrey Pines labs, Houston, TX) and anti-rabbit-555 (1:400, Jackson Immunoresearch) secondary antibody.
Hg analysis by XFI
X-ray fluorescence images were collected at the 20-ID-B beamline (PNC/XOR) at the Advanced Photon Source (Argonne, IL). The 7.0 GeV storage ring operated in continuous top-up mode at 102 mA. The incident X-ray energy was set at 13.45 keV. Experiments used a Si(111) double crystal monochromator and Rh-coated silicon mirrors for focusing and harmonic rejection. The nitrogen-filled ion chambers were used to measure the intensity of the incident X-rays. The microfocused beam of 5 μm diameter was produced by Kirkpatrick-Baez Rh-coated focusing mirrors. Samples were mounted at 45° to the incident X-ray beam and were spatially rastered in the microbeam with a step size of 10 μm. Beam exposure was 0.5 s per step. The intensities of the X-ray fluorescence lines (mercury, Hg Lα; calcium, Ca Kα; iron, Fe Kα; and zinc, Zn Kα) generated at each step were monitored using a four-element silicon-drift Vortex detector (SII NanoTechnology USA Inc.).
The XFI data were processed using SMAK software (http://home.comcast.net/∼sam_webb/smak.html). Windowed X-ray fluorescence counts were normalized by the incident X-ray intensity and background corrected by subtracting the average intensity obtained of pixels outside the tissue. Quantities of Ca, Fe, and Zn per pixel were directly calibrated by measuring the respective X-ray fluorescence calibration standard in the form of a thin layer deposited on 6.3 μm-thick Mylar film (53.3 μg/cm2 of CaF2, 51.8 μg/cm2 of Fe, and 20 μg/cm2 of ZnTe) (Micromatter, Vancouver, BC, Canada). Quantities of Hg per pixel were calibrated using two X-ray fluorescence thin standards, (1) Au—50.8 μg/cm2 and (2) TlCl—53.1 μg/cm2, deposited on 6.3 μm-thick Mylar film (Micromatter, Vancouver, BC, Canada). Using standards of gold and thallium, elements adjacent to mercury in the periodic table, was preferable to employing a mercury amalgam standard, because the latter decreased in mercury content slowly over time, presumably due to loss of elemental mercury vapor. Average background intensities for windowed fluorescence from the standards were estimated from the X-ray fluorescence image of the 6.3 μm-thick Mylar film. The background-corrected Au and Tl Lα fluorescence intensities were used to interpolate a Hg Lα fluorescence intensity, which was applied to the background-corrected Hg distribution maps to obtain the quantities of Hg per pixel in μg/cm2.
qRT-PCR for dMRP gene expression
For dMRP expression in mutant lines RNA was extracted from untreated third instar wandering larvae (approximately day 5 of larval growth). For dMRP expression in RNAi experiments, RNA was isolated from either gut tissue of adult flies (Fig. 4F) or from fat body tissue of third instar wandering larvae (Fig. 4G). Whole larval RNA was extracted from a pooled sample of ≥15 larvae disrupted in Trizol reagent (Invitrogen) using a Kontes Pellet Pestle cordless motor (Fisher no. NC0493674). Alternatively, isolated tissues from 30 flies were dissected and pooled for Trizol extraction. qRT-PCR was performed using iScript One-Step RT-PCR Kit with SYBR Green (Bio-Rad) according to manufacturer's protocol. Forty nanograms of RNA were used for each sample, and samples were run in duplicate. Samples were run on a BioRad CFX Connect Real-time System (Biorad). Fold change in expression was calculated, using either glyceraldehyde 3-phosphate dehydrogenase (GAPDH1) or ribosomal protein 49 (RP49) as a housekeeping gene, by the ΔΔCT method (Livak and Schmittgen, 2001). The following primers were used, listed forward/reverse (5’-3’): GAPDH1- TGAAGGGAATCCTGGGCTAC/ACCGAACTCGTTGTCGTACC, dMRP- ACTTTACGCCCTGCTTTGAG/TCACGTTCAGCTTGTTCCAC, and RP49-AGTATCTGATGCCCAACATCG/TTCCGACCAGGTTACAAGAAC.
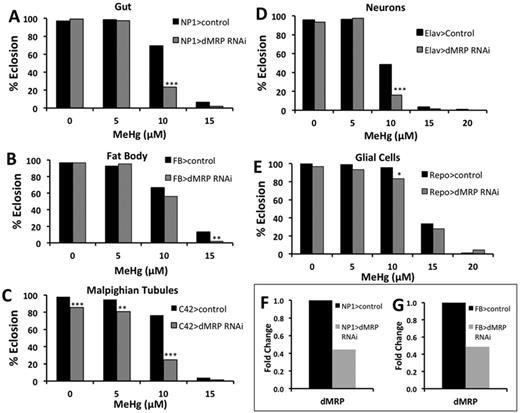
Effects of reduced dMRP expression in gut, Malpighian tubules, fat body, and nervous system on developmental toxicity of MeHg. Eclosion assays were used to determine the tolerance to MeHg during development in flies with knockdown of dMRP in various tissues. Knockdown of dMRP using UAS-dMRP RNAi was directed to the gut (A, NP1-GAL4), fat body (B, FB-GAL4), Malpighian Tubules (C, C42-GAL4), neurons (D, Elav-GAL4), and glia (E, Repo-GAL4). Asterisks indicate significant difference between dMRP knockdown and control at each treatment, as determined by z-test (*p<0.01, **p<0.001, ***p<0.0001). (F, G) Relative expression levels of dMRP subsequent to RNAi knockdown, determined by qRT-PCR with RNA extracted from gut (F) and fat body (G) tissues of either adults or third instar larvae.
Induction of dMRP upon treatment with MeHg was measured using the same methods as above. However, first instar larvae were placed on plates containing 0, 5, or 10μM MeHg for 48 h. Then, RNA was extracted from a pooled sample of 20 whole larvae.
Mercury determinations
Total Hg levels were measured in larvae of both the MRPEY11919 line and the control (y1w67c23) line using a DMA80 Direct Mercury Analyzer. First instar larvae were loaded onto plates of food with 1, 2, or 5μM MeHg and incubated at 25°C for 48 h. Larvae were then rinsed in PBS and placed on normal fly food for 4 h to clear the lumen of the gut of MeHg. Twenty-five larvae were taken from each treatment, rinsed in cold (4°C) PBS, blotted dry on filter paper, and placed into microfuge tube and frozen at −80°C for 20 min to kill the larvae. Larvae were then transferred to a nickel sample boat and weighed directly on a microbalance. Hg measurements were made in the DMA80 analyzer to determine Hg content on a weight basis expressed in μg/g (ppm). Hg determinations following this protocol were done in duplicate, separated by a one month period.
RESULTS
Localization of Hg Following MeHg Exposure in the Developing Drosophila Larva
As a first step toward characterizing dMRP function in MeHg toxicity, we sought to determine the relevant tissues targeted by MeHg in the developing fly larvae to (1) understand the relevance of the fly model to higher organisms with respect to MeHg accumulation and (2) guide our rationale for GAL4>UAS targeted expression of dMRP related transgenes. Synchrotron-based XFI is a powerful tool for visualizing the distribution of elements of interest in biological specimens down to the cellular level. Any element (sulfur and heavier) can be detected, and the presence of other elements does not decrease its efficacy. The elemental distribution images are built by raster scanning a specimen in the synchrotron micro-beam and collecting the excited X-ray fluorescence signal from each irradiated spot. The remarkably high elemental specificity of this technique lies in the fact that X-ray fluorescence photons emitted by a certain element will have a unique range of energies that cannot be mistaken with any of the other elements. XFI was used previously to study tissue-specific accumulation of Hg in MeHg-exposed intact zebrafish larvae (Korbas et al., 2008) as well as in their thin sections (Korbas et al., 2012). We therefore performed XFI analysis on larvae treated with MeHg to determine the localization of Hg in developing larval organs. For this analysis, larvae were fed on MeHg food (15μM MeHg) for a period of 72 h, followed by a recovery on control food for 4 h to clear the digestive tract of MeHg food. Paraffin sections prepared from these larvae were analyzed by XFI. The anatomy of the larvae is shown schematically in Figure 1B, with the corresponding tissues in a paraffin section labeled in Figure 1A. Elemental analysis to localize Hg, as well as zinc (Zn), iron (Fe), and calcium (Ca), was performed with XFI using a beam setting with 10 μm resolution (see Materials and Methods). Mercury is seen to be elevated in several tissues with the highest levels observed in the midgut, Malpighian tubules (renal organ), proventriculus and gastric caeca, the brain and ventral nerve cord, and the salivary gland (Fig. 1C). To a lesser extent Hg is seen in remnants of the fat body (hepatic organ); however, this tissue was not well resolved in our paraffin preparations. In addition, the muscle, associated with the cuticle layers of the larvae, showed lower but appreciable levels of Hg (Fig. 1C). An absence of Hg signal is observed in samples of larvae where MeHg was omitted from the food (Supplementary Data). The profile of Zn localization is remarkably similar to that of Hg (Figs. 1E and C). In this preparation Zn was not supplemented during feeding and is present in the standard food composition (note the Zn signal in food in the lumen of the gut compared with the absence of Hg signal, Figs. 1E and C). Also notable is the apparent higher signal for Hg in the brain and ventral nerve cord relative to Zn, indicating a preference for Hg to accumulate in this tissue (Figs. 1C and E). Iron and calcium show a more uniform distribution across all tissues relative to Hg and Zn, consistent with the ubiquitous distribution of these elements (Figs. 1D and F). However, Fe and Ca are seen to localize to distinct regions of the brain and ventral nerve cord, corresponding to the neuropil and cortex, respectively (Figs. 2C and D), indicating that specific brain regions display affinity for different classes of metal ions.
A closer examination of Hg in the brain and ventral nerve cord is presented in Figure 2, along with immunostaining of the adjacent paraffin section for the Elav antigen, which positively identifies neuronal nuclei in the cellular cortical brain regions (Figs. 2A and B). The anti-Elav staining profile confirms that Hg is elevated in the medial neuropil region relative to the cellular cortex of the brain and ventral nerve cord (Figs. 2A and B), indicating a potential preference for Hg to localize to axon tracts and/or associated glia in this region of the developing central nervous system (CNS). The specificity of this distribution of Hg is emphasized by the differential distribution of Fe and Ca seen in these same brain regions (Figs. 2C and D).
These data indicate that MeHg has a propensity to accumulate in developing Drosophila tissues that are equivalent to target tissues for MeHg found in higher organisms, namely, the gut, Malpighian tubules (renal organ), and, importantly, the CNS. Hg accumulation in the fat body (hepatic organ) is at a lower level. High Hg levels are also seen in the gastric caeca and proventriculus, invertebrate-specific organs associated with the gut that facilitate breakdown of food particles, nutrient absorption, and filtration of waste from the hemolymph.
MeHg Susceptibility in a dMRP Mutant
To assess the role of dMRP in mediating MeHg toxicity during development, we first examined two mutant fly strains carrying a disrupting insert in the dMRP gene: MRPEY11919 and MRPMI00447. In comparison to a control strain (y1w67c23) both MRP mutants demonstrate more susceptibility to MeHg in the eclosion assay, specifically on 10μM MeHg food (Fig. 3C). The MRPEY11919 mutant flies are seen to be the most sensitive to MeHg, giving lower percent of eclosion on 5μM MeHg food than the MRPMI00447 and the control line (Fig. 3C). Consistent with the increased susceptibility to MeHg, both mutant lines show decreased expression of dMRP transcript as determined by qRT-PCR, with both MRPMI00447 and MRPEY11919 showing approximately 40% expression level compared with the control (Fig. 3D). The partial depletion in dMRP expression in the MRPMI00447 line is consistent with its predicted disruption of only eight of the 17 MRP transcripts (Type A transcripts, Fig. 3B). On the other hand, the MRPEY11919 mutant unexpectedly demonstrates detectable transcript by qRT-PCR, despite the prediction that this carries a P-element insert that disrupts all the dMRP transcripts (Type A and C, Fig. 3B). The nature of the transcript resulting in this PCR amplification product is uncertain, but may indicate the presence of yet to be described RNA splice variant of dMRP that excludes the region of this P-element insert. Nonetheless, both mutants show much reduced transcript levels.
The MiMIC construct in the MRPMI00447 line, by design, carries a GFP open reading frame with associated TATA and 3’ poly A tail sequence such that, when inserted in plus orientation downstream of enhancer or regulatory regions of the gene it disrupts, it will express GFP in the pattern of that gene's expression (Fig. 3B). Analysis of GFP in living larvae of the MRPMI00447 line was therefore used to localize endogenous expression of dMRP. Examination of thin sections of MRPMI00447 larvae showed GFP expression in the midgut and Malpighian tubules (Fig. 3E). In addition, GFP was seen localized to the body wall muscle adjacent to the cuticle (Figs. 3G and H). This pattern is consistent with transcript expression analysis reported by FlyAtlas resource on the Flybase (Gelbart and Emmert, 2013). Furthermore, these data demonstrate that dMRP is expressed in two of the tissues that show highest levels of Hg accumulation by XFI analysis: gut and Malpighian tubules.
Effects of dMRP Knockdown on Development of MeHg Exposed Larvae
Based on the above XFI findings; and on established roles of the gut, Malpighian tubules, and fat body in transport, metabolism, and excretion of toxins; we chose to knockdown expression of dMRP exclusively in these tissues to assess their contribution to moderating MeHg toxicity. Tissue-specific GAL4 driver lines [NP1-GAL4 (gut), C42-GAL4 (Malpighian tubules), and FB-GAL4 (fat body), respectively] were crossed with a UAS-dMRP-RNAi responder line to diminish transcript expression specifically in these organs. Knocking down dMRP in the gut is seen to greatly enhance susceptibility of developing flies to MeHg, notably at the 10μM concentration, as can be seen by a decrease in percent eclosion relative to control (Fig. 4A). A similarly robust effect was seen with knockdown of dMRP in the Malpighian tubules (Fig. 4C). In contrast, dMRP knockdown in the fat body showed only a slight change in tolerance at 15μM MeHg in developing larvae (Fig. 4B). The efficacy of knockdown of dMRP by this RNAi approach was confirmed by qRT-PCR determination, on RNA isolated from gut and fat body tissues, which showed a 50% or more knockdown of expression (Figs. 4F and G). These data demonstrate a specific activity for dMRP in the gut and Malpighian tubules that can affect overall MeHg tolerance in the developing fly.
Because MeHg is a well-known neurotoxin, we also chose to use both a neuronal and a glial driver (Elav-GAL4 and Repo-GAL4, respectively) to knockdown expression of dMRP in the nervous system. Knockdown of dMRP in neurons (Elav>dMRP RNAi) showed an increased susceptibility to MeHg, as demonstrated by the lower percent eclosion at 10μM MeHg relative to control (Fig. 4D). When dMRP was knocked down in glia (Repo>dMRP RNAi), only a slight increase in susceptibility was seen at 10μM MeHg food concentration compared with control (Fig. 4E). In this latter analysis, it was noted that lines carrying the Repo-GAL4 driver exhibited an overall higher MeHg tolerance relative to other driver lines, likely reflecting a different genetic background of the Repo-GAL4 line. Importantly, these data indicate that dMRP activity solely in the nervous system is sufficient to influence overall developmental susceptibility to MeHg. These data also suggest that dMRP activity in neurons versus glia is more crucial for alleviating MeHg toxicity.
The results of both the RNAi system used to knockdown dMRP in a tissue-specific manner and that of the mutant dMRP lines indicate that dMRP is a critical mediator of MeHg toxicity during development. Our results also demonstrate that this effect has elements of tissue specificity, with dMRP knockdown in the gut, Malpighian tubules, and neurons having the greatest effect on MeHg tolerance during development.
Accumulation of MeHg in dMRP Mutant Larvae
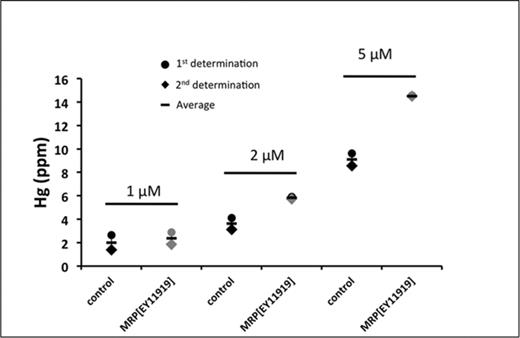
To test the possibility that increased MeHg toxicity results from elevated MeHg accumulation in dMRP mutant larvae, we fed the MRPEY11919 mutant larvae and a relevant control strain (y1w67c23) various concentrations of MeHg for 48 h. Larvae were then harvested and analyzed directly for total mercury content. At concentrations of 2μM and 5μM MeHg feeding, the MRPEY11919 mutant showed higher levels of Hg compared with the control strain. As much as 50% higher Hg levels were seen with 5μM MeHg food in the MRPEY11919 (14.5 ppm Hg) compared with the control (y1w67c23) (9.6 ppm Hg) (Fig. 5). These data suggest that diminished dMRP activity can influence the accumulation on MeHg, presumably due to compromised elimination of the toxin.
Hg retention in mutant dMRP larvae. Total Hg levels in the MRPEY11919 line and a control line subsequent to a 48-h exposure to food containing either 1, 2, or 5μM of MeHg followed by 4 h on standard food to clear the gut of all food containing MeHg. Treatments and Hg analyses were done twice (represented by circle and diamonds) and the average for the two determinations indicated by the horizontal bar. For each determination 25 larvae were pooled and analyzed for total Hg.
Effects of MeHg on dMRP Expression in Wild Strains
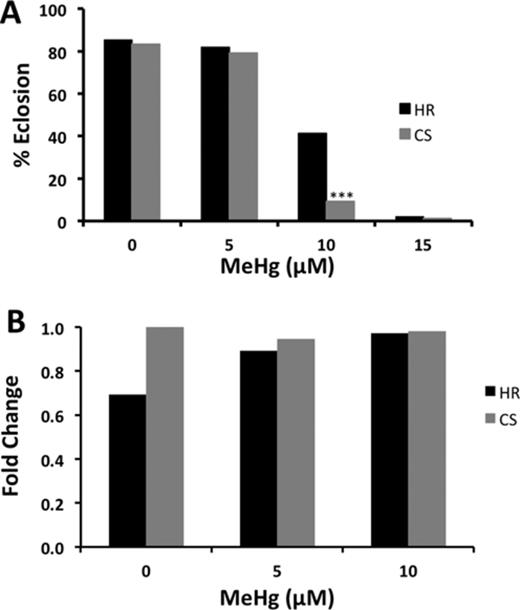
Induction of MRP transporters in response to xenobiotics has been demonstrated in several instances (Maher et al., 2005,2008). We therefore tested whether or not MeHg exposure could induce dMRP expression in two wild-type strains of flies, Canton S and Hikone R, which have previously been characterized for their low and high MeHg tolerance, respectively (Rand et al., 2012). Larvae of these two strains were treated with various concentrations of MeHg for 48 h, and total RNA was isolated for qRT-PCR determination of dMRP expression levels. Differences in tolerance in these two strains can be seen in Figure 6A, which demonstrates the higher tolerance of the Hikone R strain on 10μM MeHg. Treatment with MeHg did not induce dMRP expression in Canton S; however, expression increased moderately with MeHg treatment in Hikone R (Fig. 6B). It is of note that the basal expression dMRP in the more tolerant Hikone R strain is initially somewhat lower than the Canton S strain.
Tolerance and dMRP expression in two wild-type fly strains exposed to MeHg. (A) Tolerance to MeHg in Hikone R (HR) and Canton S (CS) was determined using the eclosion assay. Asterisks indicate significant difference between the two strains as determined by z-test (***p<0.0001). (B) Relative dMRP expression in CS and HR exposed to 0, 5, or 10μM MeHg food for 48 h. Expression levels in a pooled sample of 20 larvae were determined by qRT-PCR, normalized to RP49 and expressed relative to Canton S at the 0μM MeHg exposure.
DISCUSSION
Using a Drosophila model, we demonstrate that the developmental toxicity of MeHg can be moderated significantly by dMRP. Furthermore, we identify MeHg target organs in this model by physically locating sites of Hg accumulation with XFI, which reveals intestinal, renal, and neural tissues as targets. Correspondingly, a reduction of dMRP, both globally and in these individual tissues, substantially impacts the whole organism's susceptibility to MeHg toxicity. dMRP mutant larvae are seen to accumulate Hg to higher levels than control larvae, consistent with the notion that global reduction in dMRP can regulate the overall MeHg insult by compromising excretion of the toxin. The fact that dMRP is predominantly expressed in the gut and Malpighian tubules, and that Hg from a MeHg exposure is seen to concentrate in these tissues, is entirely consistent with the strong effects of dMRP on moderating MeHg toxicity. It is also consistent with the apparent higher accumulation of Hg in a dMRP mutant. RNAi knockdown of dMRP expression in the fat body, the hepatic organ of the fly, has little effect on overall MeHg toxicity in our assay, suggesting that unlike MRPs in mammalian liver, dMRP has less of a hepatic function for alleviating mercury toxicity. In addition, Hg is not seen to accumulate in fat body to levels seen in the gut and Malpighian tubules. One distinction is that the insect fat body, unlike the mammalian liver, is not contiguous with endothelia that perform excretion (such as the gut and Malpighian tubules); rather, it is freely suspended in the hemolymph. These fundamental differences in anatomy may explain why knockdown of dMRP in the fat body has little impact on MeHg toxicity. In contrast, a robust effect on MeHg susceptibility is seen when dMRP is knocked down exclusively in the nervous system. Under this condition it can be expected that Hg levels in the developing brain will increase and presumably enhance toxic MeHg effects on neural development. At the same time, brain-restricted knockdown of dMRP is not likely to influence overall excretion of MeHg from the organism. This finding highlights the sensitivity and specificity of nervous system development with respect to the overall development and eclosion of the fly under the stress of MeHg toxicity. Overall, the results of this study speak to the relevance of this model for investigation of underlying genetic factors that modify MeHg toxicity.
Aspects of the developing nervous system that are most affected by MeHg are not well resolved. Nonetheless, glial cells and the blood brain barrier (BBB) have been cited as critical in modulating access of MeHg to the brain (Aschner and Aschner, 1990; Kerper et al., 1992). A BBB-like function has been shown in the larval brain that is carried out by perineural glia that ensheath the optic lobes, central brain, and ventral nerve cord of the larval CNS (Mayer et al., 2009). Furthermore, a role for ABC family transporters localized to this fly BBB, specifically MDR65, the fly homolog of human MDR1/p-glycoprotein, has been documented to show orthologous function in exclusion of xenobiotics from the larval brain (Mayer et al., 2009). Localization of dMRP in the larval brain remains to be investigated in greater detail. Nonetheless, we have now localized Hg in the larval CNS with XFI, which shows a more concentrated level in the region of the neuropil of the ventral nerve cord and optic lobes relative to the cortical regions (Fig. 2A). This profile could be interpreted to suggest that regions dense in axonal projections have higher affinity for MeHg. Alternatively, the Hg could be harbored in the glia that localize to this region. Yet, our attempt to knockdown dMRP in glia (Repo>dMRP RNAi) gave little overall effect in our eclosion assay compared with dMRP knockdown targeted to neurons (Elav>dMRP RNAi). This latter result suggests a novel and preferential role for dMRP in neurons in the CNS. It is of interest that a recent characterization of a related MRP (MRP7) in a Caenorhabditis elegans model shows a specific activity in dopaminergic neurons that is able to protect against MeHg insult (Vanduyn and Nass, 2013). Future studies aimed at localizing dMRP expression in the developing Drosophila nervous system will shed light on the neural- versus glial-specific function of this important efflux transporter.
Despite the strong effects of dMRP knockdown on MeHg susceptibility, it is apparent that dMRP expression levels alone are not indicative of a fly's susceptibility to MeHg. Between the Canton S and Hikone R strains, dMRP expression levels do not correlate with MeHg susceptibility. However, dMRP expression is induced slightly with MeHg in the tolerant Hikone R strain, but remains unchanged in the Canton S, such that nearly equivalent expression levels are seen with 10μM MeHg treatment of both strains. Several genes acting independently, or in concert, are likely responsible for the overall trait of MeHg susceptibility. The induction of dMRP in the Hikone strain, although not exceeding that of Canton S, may be additive in function to an already tolerant genetic background of the Hikone R. Accordingly, we have previously observed that other metabolism genes, e.g., cytochrome p450 CYP6g1, can strongly influence MeHg tolerance in the Hikone R strain (Mahapatra et al., 2010; Rand et al., 2012). It would be of interest to explore the potential for CYP6g1 and dMRP to act in concert to give MeHg tolerance, thereby contributing to understanding the multi-genic trait of MeHg tolerance and susceptibility.
Individual variation in susceptibility to MeHg toxicity poses a concern for interpreting the risks associated with MeHg exposure. Population studies in flies have shown that there is a substantial genetic component to variation in MeHg susceptibility (Magnusson and Ramel, 1986; Mahapatra et al., 2010). Genetic analyses of human populations exposed to environmental mercury have demonstrated that polymorphisms in Phase II and Phase III metabolism genes are significantly associated with traits of mercury metabolism (Engstrom et al., 2013; Schlawicke Engstrom et al., 2008). Notably, several polymorphic variants in the human MRP2 (ABCC2) gene are shown to correlate with varied levels of urinary mercury in populations exposed to elemental mercury via small-scale gold mining (Engstrom et al., 2013). Our results in this study provide rationale for a role of MRP1 (and other MRPs) in moderating MeHg toxicity in humans. Moreover, the high conservation of dMRP with human MRPs suggests that future studies in this Drosophila model will be a means of characterizing the functional activity of clinically important human polymorphic variants in transgenic “humanized” flies.
In summary, we find that modulating expression of dMRP alters MeHg toxicity outcomes in Drosophila, consistent with a main effect of the MRP gene in moderating MeHg developmental toxicity. Increased susceptibility to MeHg during development is seen when dMRP expression is reduced, both globally and within isolated organs that are targeted by MeHg, including the nervous system. Our findings point to a specific role for dMRP in neurons in the protection against a MeHg insult. Finally, the system detailed here provides a tractable model to evaluate relevant human polymorphic variants of MRP and other genetic variants in genetic studies of mercury-exposed populations.
FUNDING
National Institute of Environmental Health Sciences at the National Institute of Health [R01 ES01219 to Philip Davidson, P.I., ViCTER Sub-award to M.D.R.)]; the University of Rochester Environmental Health Center [P30 ES001247]; the National Institute of Environmental Health Sciences Training Grant [T32 ES07026 to L.P].
We thank Robert Gordon for assistance at the 20-ID beamline at the Advanced Photon Source. PNC/XSD facilities at the Advanced Photon Source, and research at these facilities, are supported by the US Department of Energy - Basic Energy Sciences, a Major Resources Support grant from NSERC, the University of Washington, the Canadian Light Source, and the Advanced Photon Source. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02–06CH11357. We also thank Gary Myers, Edwin van Wijngaarden, and Gene Watson for critical review of the manuscript. We also thank Tanzy Love of the Department of Biostatistics and Computational Biology, University of Rochester, for assistance in statistical analyses.
REFERENCES
- diagnostic radiologic examination
- roentgen rays
- excretory function
- polymorphism
- adult
- diptera
- drosophila
- fluorescence
- food
- genes
- larva
- malpighian tubules
- membrane transport proteins
- mercury
- methylmercury compounds
- nervous system
- neurons
- neurotoxins
- synchrotrons
- diagnostic imaging
- mice
- persistence
- toxic effect
- rna interference
- genetic research
- developmental toxicity

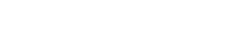


Comments