-
PDF
- Split View
-
Views
-
Cite
Cite
William T White, Shannon Corrigan, Lei Yang, Aaron C Henderson, Adam L Bazinet, David L Swofford, Gavin J P Naylor, Phylogeny of the manta and devilrays (Chondrichthyes: mobulidae), with an updated taxonomic arrangement for the family, Zoological Journal of the Linnean Society, Volume 182, Issue 1, January 2018, Pages 50–75, https://doi.org/10.1093/zoolinnean/zlx018
Close - Share Icon Share
Abstract
DNA sequence data from mitochondrial genomes and c. 1000 nuclear exons were analysed for a complete taxon sampling of manta and devilrays (Mobulidae) to estimate a current molecular phylogeny for the family. The resulting inferences were combined with morphological information to adopt an integrated approach to resolving the taxonomic arrangement of the family. The members of the genus Manta were found to consistently nest within the Mobula species and consequently the genus Manta is placed into the synonymy of Mobula. Mobula eregoodootenkee, M. japanica and M. rochebrunei were each found to be junior synonyms of M. kuhlii, M. mobular and M. hypostoma, respectively. The mitochondrial and nuclear tree topologies were in agreement except for the placement of M. tarapacana which was basal to all other mobulids in the nuclear exon analysis, but as the sister group to the M. alfredi–M. birostris–M. mobular clade in the mitochondrial genome analysis. Results from this study are used to a revise the taxonomy for the family Mobulidae. A single genus is now recognized (where there were previously two) and eight nominal species (where there were previously 11).
INTRODUCTION
The manta and devilrays of the family Mobulidae constitute some of the most charismatic species of rays. They are large (up to 7 m disc width), planktivorous species, occurring worldwide in tropical and temperate waters (Last & Stevens, 2009). Despite the public attention they receive, there is still uncertainty regarding their taxonomy, phylogeny, life history and population structure. This was highlighted by Couturier et al. (2012), whose comprehensive review of available biological and ecological data for members of the Mobulidae revealed large gaps for many parameters. In fact, the majority of studies investigating the biology and ecology of mobulid rays have focused on particular species in specific locations, limiting our ability to make generalizations at higher taxonomic levels. Very few broad regional or global studies of mobulid ray biology and ecology have been undertaken. Taxonomic research on this group has been complicated by poor representation of mobulids in biological collections because of their large size. Notarbartolo di Sciara (1987) provided a comprehensive revision of the genus Mobula, including a description of a new species. That study represented a major step forward in our understanding of mobulid taxonomy. Marshall, Compagno & Bennett (2009) revised the genus Manta, resurrecting a second species, M. alfredi (Krefft, 1868), as well as acknowledging a third putative species. Most recently, Notarbartolo di Sciara et al. (2016) provided a redescription of the poorly known Mobula kuhlii (Müller & Henle, 1841).
The Mobulidae currently comprises two genera, Manta and Mobula. The genus Manta encompasses two nominal species, the reef manta M. alfredi (Krefft, 1868) and the giant manta M. birostris (Walbaum, 1792), and possibly a third species (M. sp. cf. birostris sensu Marshall et al., 2009). There are nine currently recognized species in the genus Mobula: the pygmy devilray M. eregoodootenkee (Bleeker, 1959), the Atlantic devilray M. hypostoma (Bancroft, 1831), the spinetail devilray M. japanica (Müller & Henle, 1841), the shortfin devilray M. kuhlii, the giant devilray M. mobular (Bonnaterre, 1788), Munk’s devilray M. munkiana Notarbartolo di Sciara, 1987, the lesser Guinean devilray M. rochebrunei (Vaillant, 1879), the Chilean devilray M. tarapacana (Philippi, 1892), and the bentfin devilray M. thurstoni (Lloyd, 1908). Previous studies of mobulid phylogeny, based on both morphological (Adnet et al., 2012; Paig-Tran et al., 2013) and molecular data (Aschliman, 2011, 2014; Naylor et al., 2012b; Poortvliet et al., 2015) indicate that the family is a monophyletic lineage comprising three clades. One clade includes the larger mobulid species Manta spp., M. tarapacana, M. mobular and M. japanica. The remaining two clades comprise the smaller species M. kuhlii–M. eregoodootenkee– M. thurstoni and M. munkiana–M. rochebrunei– M. hypostoma.
Despite this progress in characterizing mobulid diversity, mobulid taxonomy overall remains largely unresolved due to a very complicated nomenclatural history and the fact that phylogenetic inferences have been limited by gaps in taxonomic and/or genomic sampling. Notable long-standing uncertainties regarding mobulid taxonomy include the validity of the genus Manta (Herman et al., 2000; Adnet et al., 2012; Paig-Tran et al., 2013; Naylor et al., 2012b; Aschliman, 2014; Poortvliet et al., 2015), as well as distinguishing species boundaries from intraspecific geographical variants within multiple lineages of Mobula. Specifically, gross morphological and/or genetic similarities have been noted between species pairs M. kuhlii/M. eregoodootenkee, M. hypostoma/M. rochebrunei and M. mobular/M. japanica that suggest they may possibly be conspecifics (Notarbartolo di Sciara, 1987; Paig-Tran et al., 2013; Poortvliet et al., 2015; Henderson et al., 2016). Phylogenetic inferences based on morphology have either been more concerned with the phylogenetic placement of mobulids within Myliobatiformes rather than the relationships among mobulids, or have focused on the evolution of particular structures. Making taxonomic decisions based on these works has thus largely been limited by incomplete taxon sampling.
Molecular phylogenetic inferences have considered a more complete taxon sampling and provided interesting insights regarding the evolutionary history of Mobulidae. However, these have been exclusively based on the mitochondrial genome (Aschliman, 2011; Naylor et al., 2012b; Poortvliet et al., 2015). Two recent estimates of mobulid phylogeny were based on data collected from both the mitochondrial and nuclear genomes; however, only a small number of nuclear markers were assessed and the data lacked resolution (Aschliman, 2014; Poortvliet et al., 2015). This is problematic, especially for the purposes of distinguishing species boundaries and making taxonomic decisions among closely related lineages. Coalescent variation and the potential for gene tree–species tree discordance (Maddison, 1997) are now well-documented and estimating phylogenies based on a small number of molecular markers is now considered insufficient for these purposes. Moreover, many factors and processes can lead to differential phylogenetic signal between the mitochondrial and nuclear genomes. These include lineage sorting, demographic asymmetries, selection and hybridization. Although taxonomic uncertainties have been discussed, all authors have favoured taxonomic stability for the group in light of their various limitations.
There is considerable concern regarding the conservation status of mobulid rays globally, evidenced by the inclusion of Manta spp. in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) in 2013 (CoP16). It is argued that Mobula spp. are subject to much higher levels of exploitation than Manta spp. in some regions (e.g. Indonesia; White et al., 2006) and these too were listed on CITES Appendix II in 2016 (CoP17). Further research that can inform the conservation management of these species is thus urgently required. Taxonomy is recognized as the foundation that underpins our knowledge of biodiversity (Simpfendorfer et al., 2011). From a conservation management perspective, it is crucially important to accurately recognize the biodiversity within vulnerable groups, such as the mobulids, so that conservation priorities may be identified and management strategies devised that are both efficient and effective. The most robust approach to identifying taxonomic boundaries is to assess multiple data types from a large number of comparative samples. However, when considering rare and endangered animals, this becomes impractical and employing the precautionary principle may mean making taxonomic decisions based on the best available information. In this paper, we address some of the limitations of previous studies to provide an accurate taxonomic accounting of all nominal species of mobulids and an estimate of their phylogenetic relationships. Phylogenetic analyses of DNA sequence data obtained using a targeted gene capture approach were carried out. The dataset comprised the protein coding components of the whole mitochondrial genome sequences and aligned sequences derived from more than 1000 putatively single-copy nuclear exons for all extant members of the family Mobulidae. This approach is an improvement over previous studies because the inference is based on a large number of loci from both the mitochondrial and nuclear genomes, reducing the probability of error associated with differences in the coalescent history among individual genes. Moreover, the particular gene capture approach used minimizes paralogous gene comparisons within the dataset a priori. Any discordance between our molecular phylogenetic inference and the previously recognized taxonomy of this group was resolved by undertaking a detailed examination of morphological data and the nomenclatural history for the taxa involved. We present a revised taxonomy for this iconic group of rays based on our results.
MATERIAL AND METHODS
Taxon sampling
Specimens examined are listed in the Material Examined section. Museum acronyms follow Fricke & Eschmeyer (2015).
Muscle tissue samples were collected for DNA analysis from a complete taxon sampling of the mobulid rays (11 described species; tissue accessions are listed in Material Examined section), and from three outgroup species (Rhinoptera bonasus, tissue accession GN5465; Myliobatis aquila, tissue accession GN7203; Aetobatus narinari, tissue accession GN5677). Tissue was stored in 95% ethanol or 20% salt-saturated DMSO prior to DNA extraction using the E.Z.N.A. Tissue DNA Kit (Omega Bio-Tek, Inc., Norcross, GA, USA), following the manufacturer’s instructions. A single set of universal primers was used to amplify the mitochondrial DNA NADH2 fragment by polymerase chain reaction (PCR) for all samples prior to purification and Sanger sequencing, as described in Naylor et al. (2012a). This fragment is particularly useful for distinguishing elasmobranch species and was used primarily to ensure that no labelling errors had occurred in the field by confirming the nominal species identification of each specimen, prior to further analysis. This was achieved by comparing the obtained sequences against a reference database curated within our laboratory that contains ~12000 elasmobranch NADH2 sequences, including many vouchered specimens (Naylor et al., 2012b). The only available tissue sample of Mobula rochebrunei originated from the dry holotype specimen (see Material Examined) and failed to yield any results. This species is thus not represented in our nuclear DNA analyses. However, the complete mitochondrial genome sequence, derived from this same holotype specimen, was available via Genbank (Accession number KM364992.1; Poortvliet et al., 2015) and was included in our analyses of mitochondrial DNA data (details below).
Data generation: library preparation, DNA hybridization capture and next-generation sequencing
Genomic DNA was quantified using a Qubit 2.0 Fluorometer (Life Technologies Corporation, California, USA). Based on an assessment of DNA quality, a single representative of each species/lineage (nominal ID confirmed via NADH2 sequence) was chosen and subjected to targeted DNA hybridization capture for the purposes of collecting complete mitochondrial genome and nuclear exon sequences. Genomic DNA (0.5–3 μg per sample) was sheared to c. 500 bp using acoustic ultrasonication on a Covaris M220 Focused-ultrasonicator (Covaris, Inc., Massachusetts, USA) to form a target library for each specimen. Illumina sequencing libraries (Meyer & Kircher, 2010) were then prepared using the ‘with-bead’ method (Fisher et al., 2011), following Li et al. (2013). Two custom biotinylated RNA bait libraries (MYbaits MYcroarray, Ann Arbor, MI, USA) were used in two separate DNA hybridization experiments per sample. The first bait library targeted the entire mitochondrial genome and included bait sequences derived from 83 batoid species. The second bait library included sequences derived from five batoid species and targeted 1088 slow-evolving, nuclear exons that were identified previously to be putatively single-copy orthologs across six available model vertebrate genomes (Homo, Anolis, Callorhinchus, Danio, Gallus and Xenopus; Li et al. 2013 for details). Both the mitochondrial and nuclear bait sequences spanned the taxonomic diversity of batoids to allow capture experiments to be performed across divergent target species using a single set of baits for each marker type. Cross-species DNA hybridization capture followed the relaxed hybridization method described by Li et al. (2013). The enriched individual sample libraries were pooled in equimolar ratios and pooled libraries were diluted to 12–15 pM for paired-end 250–300 bp sequencing on an Illumina MiSeq benchtop sequencer (Illumina, Inc., San Diego, CA).
Sequence read mapping, contig assembly and orthology testing
Sequence reads associated with each sample were identified and sorted by their respective indices Li et al. (2013). Adapters and low-quality reads were removed using cutadapt and FastQC available in the wrapper script Trim Galore! v0.3.1 (Krueger, 2012).
Trimmed mitochondrial sequence reads were imported into Geneious Pro v7.1.9 (Biomatters Ltd, Auckland, New Zealand. Available at http://www.geneious.com) and unique reads were retained and mapped to the reference sequence of a closely related species (either sequenced by us or downloaded from GenBank).
The amino acid sequences of the 1088 putatively single-copy nuclear exons were aligned across the six model vertebrates using the – auto option to MAFFT v7. 023 (Katoh & Standley, 2013). Poorly aligned sequences were removed using trimAl v1.2rev59 (Capella-Gutierrez, Silla-Martinez & Gabaldón, 2009). The orthologous group alignments were used to build profile-hidden Markov models (pHMMs; Eddy, 1998) using HMMER (Eddy, 2011) that would serve as a reference database for assigning target sequences to orthologous groups. A BLAST (Altschul et al., 1997) database was constructed from the sequences of Callorhinchus, which we used as our reference taxon (a chondrichthyan fish and therefore the most suitable reference taxon of the available model vertebrates, Venkatesh et al., 2005). De novo assembly of trimmed sequence reads resulting from the enriched target libraries was performed with ABySS v1.3.6 (Birol et al., 2009) using multiple k-mer values (k = 51 to k = 251, in increments of 10). Assembled contigs were filtered, extended and merged using Trans-ABySS v.1.4.4 (Robertson et al., 2010). HaMStR v13.2.3 (Ebersberger, Strauss & von Haeseler, 2009), which uses a combination of BLASTP (Altschul et al., 1997), GeneWise (Birney, Clamp & Durbin, 2004) and HMMER, was used to search the assembled contigs of each sample for protein sequences that matched the database of orthologous groups. Any contig that matched a pHMM with an E-value less than 1.0 × 10−5 was initially considered a ‘hit’ to that orthologous group. Hits against the pHMMs were retained only if they fulfilled a reciprocal best BLAST criterion with the reference taxon. When multiple contigs fulfilled the orthology criteria for a particular locus, the sequence with the best score in the initial pHMM search was retained as the representative for that locus.
Alignment and phylogenetic analyses
All analyses of the NADH2, mitochondrial genome and nuclear exon datasets were performed in PAUP* v4.0a148 (Swofford, 2002) unless otherwise specified.
Nucleotide sequences of the NADH2 fragment were aligned using Geneious Pro v.6.1.7. Translation of the resulting nucleotide alignment confirmed that it was in frame and free of stop codons (which may indicate sequencing errors or misalignment). The original nucleotide alignment was used for analysis and was 1044 bp in length, including 314 parsimony-informative sites (GenBank accession numbers KU999796–KU999882). In addition to being used to confirm nominal species identifications, this dataset was also used to explore intra- versus inter-specific divergences for larger sample sizes than that could be included in our DNA hybridization capture experiments. Pairwise uncorrected p-distance (the proportion of sites at which two sequences differ) was calculated for the NADH2 dataset. The TVM + I + G model was selected as the most likely model of sequence evolution using the corrected Akaike Information Criterion (−ln L = 5184.8, AICc = 10674.8; Posada & Buckley, 2004). The maximum likelihood (ML) tree was estimated using a heuristic search that applied the parameter estimates that were identified during model selection.
For ease of alignment, only the protein-coding components of the mitochondrial genomes were aligned across all taxa using Geneious Pro v7.1.9, yielding an alignment that was 11442 bp in length, including 2711 parsimony-informative sites (GenBank accession numbers KX151642–KX151654). The complementary strand sequences were used for ND6, which is encoded on the L-strand. Again, alignment quality was confirmed via translation and incomplete stop codons were excluded from the alignment. The optimal partitioning and model scheme for the original nucleotide dataset was identified as a 17-partition scheme (Supporting Information Table S1). The ML tree was estimated using a heuristic search as described previously for the NADH2 dataset but which applied the partitioning/model scheme and parameter estimates that were identified during model selection. Support values for nodes were obtained via nonparametric bootstrapping (100 replicates).
The identified putative nuclear ortholog protein sequences for each sample were back-translated to their original nucleotide sequences using a custom Perl script, aligned using MAFFT v7.02, and concatenated with a custom Perl script. This yielded a data matrix that represented 1082 exons, was 290121 bp in length and 97.4% complete. The GTR + I + G model was selected as the most likely model of sequence evolution (−lnL = 583 275.2, AICc = 1 166 616.4) and ML analyses including nonparametric bootstrapping were performed as described previously. Although our DNA hybridization capture array targets exons that are known a priori to be single-copy across vertebrates, we are not able to account for ‘inparalogs’ resulting from duplication events within Chondrichthyes (Li et al., 2013). We therefore attempted to remove any remaining potentially paralogous comparisons from our dataset by conducting a stringent likelihood ratio test that compared clock and non-clock-like models for each exon. Exons that had a P-value less than 0.05 for the likelihood ratio test of rejection of a clock model in favour of a non-clock model were excluded from analysis as potentially including paralogous sequences. Maximum likelihood analyses were performed on the concatenated filtered dataset (144261 sites from 614 exons; filtered nuclear data are archived in TreeBASE https://treebase.org; study ID S19059) in the same manner as described above (TIM + I + G model –ln L 266 500.3, AICc = 533 062.6; parameter estimates identified during model selection).
Species tree inference can be complicated by hybridization, incomplete lineage sorting (ILS) and gene duplication/loss (Maddison, 1997). Failing to account for these processes can sometimes yield incorrect estimates of the species tree (Roch & Steel, 2015). Incomplete lineage sorting is the most commonly purported cause of gene tree–species tree discordance and is typically modelled by the multi-species coalescent (Kingman, 1982). While this approach is robust to the distorting effects of ILS, it has been shown that concatenated analyses can often be more effective than multi-species coalescent approaches when the level of ILS is low (Chou et al., 2015). Because we did not know the extent of ILS in the dataset a priori, we employed both a maximum likelihood analysis of the concatenated nuclear dataset and a multi-species coalescent-based approach (SVD Quartets: Chifman & Kubatko, 2014). Analyses were conducted on both the complete nuclear exon dataset and the subset of clock-like exons.
RESULTS
Phylogenetic analyses
Nominal species identifications for all samples as well as the relationships between them based on an ML analysis of their mitochondrial NADH2 sequences can be seen in Supporting Information Fig. S1. Intraspecific divergences based on the NADH2 marker and uncorrected p-distance measure were low, ranging between 0 and 0.011, with an average of 0.003. Notably, interspecific comparisons of p-distance between samples nominally identified as Manta birostris versus Manta alfredi (range 0–0.012, average 0.007), M. kuhlii versus M. eregoodootenkee (range 0.000–0.008, average 0.005), M. mobular versus M. japanica (range 0.004–0.011, average 0.005) and M. rochebrunei versus M. hypostoma (range 0.001–0.003, average 0.002) were of the same magnitude as observed intraspecific divergences in closely related taxa. This is reflected in the very close relationships resolved between these species pairs based on the ML analysis of the NADH2 data. Manta birostris and M. alfredi form a single closely related clade, as do M. kuhlii and M. eregoodootenkee, M. mobular and M. japanica, and M. rochebrunei and M. hypostoma (Supporting Information Fig. S1). Excluding these particular interspecific comparisons, the range of all other interspecific divergences was 0.026–0.148, with an average of 0.123.
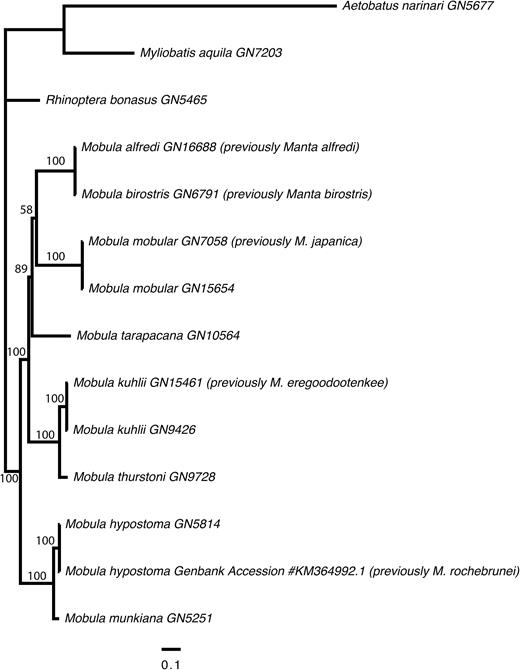
Pairwise sequence divergence based on protein coding mitochondrial genome sequences ranged from 0.001 to 0.116. Consistent with the results based on the NADH2 data, comparisons falling at the lower end of this range were those between M. birostris and M. alfredi (p-distance = 0.004), M. kuhlii and M. eregoodootenkee (p-distance = 0.005), M. mobular and M. japanica (p-distance = 0.002) and M. rochebrunei and M. hypostoma (p-distance = 0.001). Uncorrected p-distance based on the mitochondrial genomes for all other pairwise comparisons were at least an order of magnitude higher. The ML tree inferred from the mitochondrial genome data is well resolved into three major clades, largely with good support. Notably, the two Manta species are nested within Mobula, forming a sister relationship with Mobula mobular (including the specimen nominally assigned as M. japanica, Fig. 1). This grouping is sister to M. tarapacana. It should be noted that the relationships within this clade were resolved with somewhat lower bootstrap support than other relationships in the tree and thus should be interpreted accordingly (Fig. 1). A sister relationship was resolved between M. kuhlii (including the specimen nominally assigned as M. eregoodootenkee) and M. thurstoni with 100% bootstrap support. Finally, M. munkiana is sister to M. hypostoma (including the specimen nominally assigned as M. rochebrunei), also with 100% bootstrap support. This clade is basal to a sister relationship between the M. tarapacana–Manta–M. mobular clade and the M. kuhlii–M. thurstoni clade.
Phylogenetic tree showing the relationships among mobulid species, relative to three outgroups (Aetobatus narinari, Myliobatis aquila and Rhinoptera bonasus). The tree was derived from a Maximum Likelihood analysis of an alignment of the protein coding components of the mitochondrial genomes (11442 sites) under a partitioned model of molecular evolution. Bootstrap support values are displayed on the nodes.
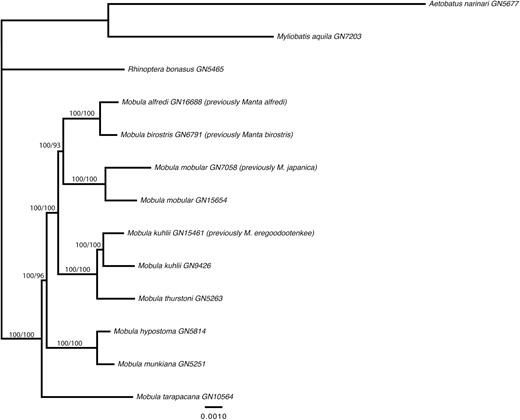
Pairwise uncorrected p-distance based on the nuclear data was much lower than that observed for the mitochondrial data, ranging from 0.002 to 0.021. Again, comparisons involving M. birostris and M. alfredi (p-distance = 0.003), M. kuhlii and M. eregoodootenkee (p-distance = 0.006) and M. mobular and M. japanica (p-distance = 0.005) were at the lower end of this spectrum. Identical tree topologies were resolved from the concatenated ML and SVD Quartets analyses of both the complete and clock-filtered nuclear exon datasets; thus we depict only the tree resulting from the ML analysis of the filtered data. Support values are shown for the concatenated ML and SVD Quartet analyses (Fig. 2). The tree topology was well resolved, with good support and topologically similar to that obtained from analyses of the whole mitogenome data. The two Manta species are again nested with Mobula, sister to M. mobular (including the specimens nominally assigned as M. japanica) with 100% and 93% bootstrap support for the filtered concatenated ML and SVD Quartet analyses, respectively. Mobula kuhlii (including the specimens nominally assigned as M. eregoodootenkee) falls as sister to M. thurstoni (100% bootstrap support); and M. hypostoma as sister to M. munkiana (100% bootstrap support). The major difference between the tree topologies derived from the nuclear and mitochondrial analyses concerns the placement of M. tarapacana. In the nuclear data, M. tarapacana falls basal to all other mobulid lineages with 100% and 96% bootstrap support for the filtered concatenated ML and SVD Quartet analyses, respectively, whereas it forms a clade with M. birostris, M. alfredi and M. mobular in the mitochondrial analyses.
Phylogenetic tree showing the relationships among mobulid species, relative to three outgroups (Aetobatus narinari, Myliobatis aquila and Rhinoptera bonasus). The tree was derived from an ML analysis of a concatenated alignment of 614 ‘clock-like’ exons (144261 sites) under a TIM + I + G model of molecular evolution. Bootstrap support values are displayed on the nodes for concatenated ML and SVD Quartet analyses.
TAXONOMIC IMPLICATIONS
Genus Mobula Rafinesque, 1810
Mobula Rafinesque, 1810, 48, 61 (type species Mobula auriculata Rafinesque, 1810; by monotypy)
Apterurus Rafinesque, 1810: 48, 62 (type species Apterurus fabronii Rafinesque, 1810; by monotypy)
Cephalopterus Risso, 1810: 14 (type species Raja giorna Lacepède, 1803; by subsequent designation)
Apturus Rafinesque, 1815: 93 (emended spelling for Apterurus Rafinesque, 1810)
Cephaloptera Cuvier, 1816: 138 (type species Raja cephaloptera Bloch & Schneider, 1801; by absolute tautonymy)
Dicerobatus Blainville, 1816: 112 (type species Raia mobular Bonnaterre, 1788; by monotypy)
Dicerobatis Blainville, 1825: 40 (type species R. mobular Bonnaterre, 1788; unjustified emendation of Dicerobatus Blainville)
Manta Bancroft, 1829: 454 (type species Cepha lopterus manta Bancroft, 1829; by monotypy)
Cephalopteram Griffith & Smith, 1834: 617 (erroneous spelling for Cephaloptera Cuvier, 1816)
Ceratoptera Müller & Henle, 1837: 118 (type species Cephaloptera giorna Lesueur, 1824; by subsequent monotypy)
Brachioptilon Hamilton in Newman, 1849: 2358 (type species Brachioptilon hamiltoni Hamilton, 1849; by monotypy)
Diabolicthys Holmes, 1856: 45 (type species Diabolicthys elliotti Holmes, 1856; by monotypy)
Deratoptera Krefft, 1868: 3, 9, Fig. (considered a misspelling of Ceratoptera Müller & Henle, 1837)
Ceratobatis Boulenger, 1897: 227 (type species Ceratobatis robertsii Boulenger, 1897; by monotypy)
Pterocephalus Swainson, 1838: 170, 174 (type species R. giorna Lacepède, 1803; replacement name)
Pterocephala Swainson, 1839: 321 (alternative spelling of Pterocephalus Swainson, 1838)
Daemomanta Whitley, 1932: 327 (type species Ceratoptera alfredi Krefft, 1868; by original designation)
Indomanta Whitley, 1936: 11 (type species Indomanta tombazii Whitley, 1936; by monotypy)
Definition: (Adapted from Bigelow & Schroeder, 1953). Medium to very large rays with a rhomboidal disc, much wider than long, depressed and relatively thick. Head broad, dorsally flattened, protruding forward anteriorly of eye level; prominent cephalic lobes extending forward on each side of head, widely separated, curving forward from front of head; eyes positioned laterally on head; spiracles subcircular to slit-like, located either dorsal or ventral to plane of pectoral disc; mouth very broad, nearly straight, located either terminally or subterminally on head; numerous minute teeth present in tooth bands in either both jaws, or only in the lower jaw. Tail long to relatively short, whip-like, usually less than width of disc; small dorsal fin present opposite pelvic-fin bases; a small, serrated stinging spine sometimes present. Skin either naked or rough with numerous small denticles. Gill arches with numerous gill plates (50 to at least 140); gill plates thin, membranous or somewhat horny with cartilaginous basal supports; outer edge of gill plates with lateral lobes that are either rounded and separated from those on adjacent plates, or rod-like and fused to those on adjacent plates.
Nomenclatural discussion: In addition to the generic synonyms listed above, Lacepède (1798) named the genus Aodon for Squalus massasa, Squalus kumal and Aodon cornu for species lacking teeth. Squalus massasa and S. kumal were named by Forsskål (1775) from the Red Sea, but no adequate description is available to allow for species determination. Squalus massasa is considered a problematic species that was described as having long pectoral fins but no teeth. Aodon cornu was an unneeded new name for Squalus edentulus Brünnich, 1768, which equals M. mobular (Bonnaterre, 1788). Aodon has therefore been used for a Mobula species, predating Mobula Rafinesque, 1810; however, the type species for this genus was subsequently designated by Jordan & Evermann (1896) as S. massasa.
Remarks: Separation of the genera Manta and Mobula has long been upheld due to the striking feature of a terminal versus a subterminal mouth, respectively. However, the comprehensive genetic analyses undertaken in this study provide the strongest evidence to date that separation of these genera is not warranted. The species previously designated to Manta, M. alfredi and M. birostris are clearly nested within Mobula, forming a close relationship with M. mobular and perhaps M. tarapacana, based on independent analyses of both mitochondrial genomes (Fig. 1) and more than 1000 nuclear exons (Fig. 2). Although a terminal versus subterminal mouth is a strong character, the dorsal versus ventral position of the spiracle in mobulids is another significant character. Both M. alfredi and M. birostris have slit-like spiracles located dorsal to the plane of the pectoral fins similar to those of M. mobular and M. tarapacana, and different to the subcircular spiracles located ventral to the plane of the pectoral fins in the remaining smaller Mobula species. This morphological character, therefore, largely supports the finding based on analysis of the mitochondrial genomes that M. birostris and M. alfredi form a clade with M. mobular and M. tarapacana, to the exclusion of the smaller mobulid species (Fig. 1). Both M. alfredi and M. birostris are also inferred as sister to M. mobular based on the independent analysis of nuclear exon data (Fig. 2), providing further support that the genus Manta is invalid. It should be noted, however, that a slightly different topology that places M. tarapacana basal to all other mobulid species is consistently resolved by the nuclear analysis with strong support (Fig. 2). This highlights the importance of integrated approaches to resolving taxonomy and inferring phylogeny, using both molecular and morphological approaches in combination.
Mobula hypostoma (Bancroft, 1831)
Cephalopterus hypostomus Bancroft, 1831: 134 (Jamaica; holotype not preserved)
Cephaloptera olfersii Müller, 1836: 311 (Brazil; syntypes MNHN A-9966,?ZMB 31636 [ex ZMB 8923],?ZM 31637)
Cephaloptera massenoidea Hill, 1862: 176 (Jamaica; no types known)
Cephaloptera rochebrunei Vaillant, 1879: 187 (Senegal; MNHN A-9967)
Ceratobatis robertsii Boulenger, 1897: 227 (Jamaica; holotype BMNH 1897.7.1.40)
Common name: Atlantic devilray
Nomenclatural discussion: Bancroft (1831) designated a new species name, C. hypostomus, to a devilray specimen from Jamaica. He only distinguished this new species from his C. manta Bancroft, 1829 (= Mobula birostris) in the form of the anterior margin of pectoral fins, a ventrally positioned mouth, and a rounded spiracle located on ventral plane and not dorsal plane of disc. Although the specimen is mentioned to have been included in the collection, there is no mention of the type being preserved. No neotype was designated by Notarbartolo di Sciara (1987) as there was no species identification issues with this species in the western Atlantic.
In 1836, Müller designated the name C. olfersii for a Brazilian species based on a skeleton and head (ZMB syntypes) and a dry specimen (MNHN specimen). This has been considered a junior synonym of M. hypostoma (Bigelow & Schroeder, 1953; Notarbartolo di Sciara, 1987). Two other new combinations were designated for Jamaican material in subsequent decades, that is, C. massenoidea Hill, 1862 and C. robertsii Boulenger, 1897, with both being junior synonyms of M. hypostoma.
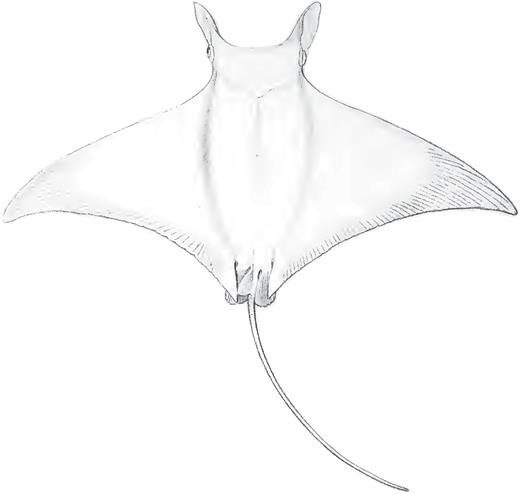
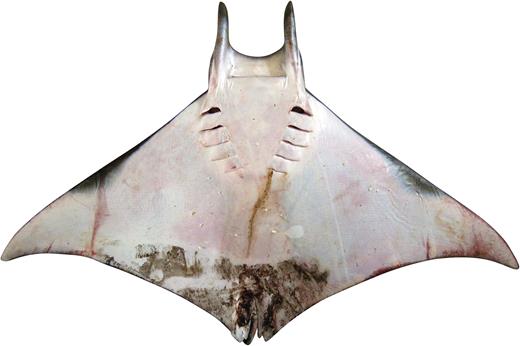
Vaillant (1879) described C. rochebrunei from a single specimen from Senegal in the Eastern Atlantic (Fig. 3). Bigelow & Schroeder (1953) considered this species a synonym of M. hypostoma and included Senegal in the range of this species. In contrast, Krefft & Stehmann (1973) listed it as a synonym of M. mobular. Notarbartolo di Sciara (1987) concluded that M. rochebrunei was distinct from M. hypostoma based on newly acquired morphometric data for several characters, for example, distance between first gill slits and predorsal length, dentition (tooth crown crenulated on labial edge vs. smooth).
Dorsal view of the dry, stuffed holotype of Mobula rochebrunei: MNHN A9967, adult male 108.5 cm DW.
Remarks: This study provides a substantial amount of molecular data allowing a direct comparison of M. rochebrunei to M. hypostoma. Analysis of the mitochondrial genome data shows an extremely close relationship between the holotype of M. rochebrunei and M. hypostoma (Fig. 1). The observed uncorrected p-distance between these taxa was the lowest of all pairwise comparisons within Mobulidae, and likely within the realm of representing intraspecific variation. Thus, based on these results, we conclude that M. rochebrunei is a junior synonym of M. hypostoma, with the latter species confirmed as occurring in both the Western and Eastern Atlantic. The morphometric and dentition differences highlighted by Notarbartolo di Sciara (1987) were based on low sample sizes. The differences seen in several characters most likely represent intraspecific variation. A more detailed taxonomic review of the Eastern versus Western Atlantic M. hypostoma populations is required to elucidate whether there are population-level differences, or whether those differences simply reflect the low sample size available.
Mobula kuhlii (Müller & Henle, 1841)
Cephaloptera kuhlii Müller & Henle, 1841: 185, Pl. 59 (left) (India; lectotype MNHN 000-1596)
Dicerobatis eregoodoo Cantor, 1849: 1420 (Penang, Malaysia and Coromandel, India; syntype location unknown)
Dicerobatis draco Günther, 1872: 422 (Misol, Indonesia; syntypes BMNH 1870.8.31.68–69)
Common name: Pygmy devilray
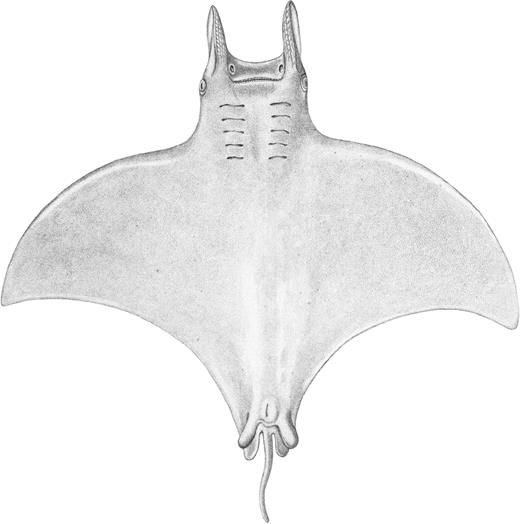
Nomenclatural discussion: The description of M. kuhlii by Müller & Henle (1841) does not provide adequate information to determine the identity of this Mobula species (Fig. 4), but examination of the lectotype (Fig. 5) and paralectotype enables it to be clearly distinguished from M. japanica, M. tarapacana and M. thurstoni. Mobula eregoodootenkee has a very complicated nomenclatural history, which is still largely unresolved. Russell (1803) provided a basic illustration and limited description of a small mobulid caught off south-eastern India, locally known as ‘Eregoodoo tenkee’ (Fig. 6). Although the description of the 4 ft. 5 in. (~135 cm) DW specimen was brief, it did include the following diagnostic characters: no spine on tail, spiracles absent behind the eyes (suggesting that they were ventral to plane of disc in the specimen examined), narrow strip of granular teeth in each jaw, and mouth behind front of head (not terminal; see Fig. 6). These characters, together with the capture location of India, indicate that the species Russell examined was one of the small Indo-West Pacific species with spiracles below the plane of pectoral disc, that is, M. eregoodootenkee, M. kuhlii or M. thurstoni. However, Russell did not designate a type and did not name it binomially; thus, as with other species described in the same publication, Russell’s ‘Eregoodoo tenkee’ is not an available name. In Cuvier (1829), ‘Eregoodoo-tenkee, Russ., I, 9’ is listed in a footnote on page 402 and some authors considered him to be the authority for this species, as Cephaloptera eregoodoo-tenkee Cuvier, 1829 (Notarbartolo di Sciara, 1987). However, Cuvier (1829) did not allocate this species to Cephaloptera; so, this combination is not valid. Also, when compared to the style used elsewhere in Cuvier (1829), it appears to be presented in the vernacular and thus not an available binomial name.
Original illustration of Cephaloptera kuhlii in Müller & Henle (1841).
Dorsal view of the alcohol-preserved lectotype of Mobula kuhlii (MNHN 000-1596, juvenile male 71.7 cm DW).
Original illustration (ventral view) of Eregoodoo tenkee in Russell (1803); presented only as a vernacular name thus not an available name.
The authority for this species has most recently been considered to be Bleeker (1859) with this authority considered the first proper binomial name attributed to this species. Bleeker lists this species as: ‘Cephaloptera eregoodoo tenkee Cuv. = Eregoodoo tenkee = Russ., fig. 9, 10 = Cephaloptera Olfeisii J. Mull. = Indian Cephaloptera J. E. Gray = Dicerobatis eregoodoo Cant., Cat. Mal. Fish. p. 438’. From this information, it is clear that Bleeker is referring to Cuvier’s use of Russell’s ‘Eregoodoo tenkee’. No descriptive features are provided and thus the identity of this species is still not determinable. Two other binomial names are also presented in Bleeker (1859). The first, Cephaloptera Olfeisii, equals C. olfersii Müller, 1836, which is a junior synonym of M. hypostoma (Bancroft, 1831) occurring in the Western Atlantic. Müller & Henle (1841) included Russell’s Eregoodoo Tenke in the synonymy of C. olfersii although the only distribution provided was Brazil. The second is D. eregoodoo Cantor, 1849, which is presented as a new name combination for Cuvier’s species, that is, D. eregoodoo (Cuvier), from Coromandel in India and Penang in Malaysia. Cantor provided a detailed description of this species, including morphometrics, based on a young male specimen (~78 cm DW) from Penang on the west coast of peninsular Malaysia. In this description, Cantor states that it agrees with Russell’s ‘Eregoodoo Tenke, No. IX. R, (not No. IX, N, from St. Helena)’ in several characters, but clearly states that the only individual examined is the Penang specimen. Unfortunately, the whereabouts of the syntype(s?) is unknown. Although most of the descriptive characters are generic for most Mobula species, several key features are provided. For example, the location of the spiracles beneath the origin of the pectoral fins discounts the two large Indo-Pacific species, M. japanica (Müller & Henle, 1841) and M. tarapacana (Philippi, 1892), which have the spiracles located above the pectoral-fin origins. Also, the teeth are described as being twice as wide as long, which is similar to that described and illustrated for this species in Notarbartolo di Sciara (1987). In contrast, M. thurstoni (Lloyd, 1908) was found to have longer teeth, not twice as wide as long. Thus, of the currently known Indo-Pacific species, M. kuhlii (Müller & Henle, 1841) and M. eregoodootenkee (sensu Notarbartolo di Sciara, 1987) are the only options left for Cantor’s D. eregoodoo. Notarbartolo di Sciara (1987) considered D. eregoodoo Cantor, 1849, to most likely be M. thurstoni due to the white spot on the apex of the dorsal fin, but this character has been found to be variable in at least one species, that is, M. kuhlii. If considered a valid species, Mobula eregoodoo (Cantor, 1849) would be the correct name for this taxon, not M. eregoodootenkee (Cuvier, 1829) or M. eregoodootenkee (Bleeker, 1859). The latter two combinations should be considered nomen dubium since they provide no characters to distinguish which taxon they denote.
The distinction between M. kuhlii and M. eregoodootenkee (sensuNotarbartolo di Sciara, 1987) has previously been based almost entirely on the length of the cephalic lobes, with the latter species having very long cephalic lobes (> 16% DW) versus relatively short in M. kuhlii (< 15% DW). The neotype of M. eregoodootenkee designated by Notarbartolo di Sciara (CAS 56095) illustrates this feature well (fig. 11 in Notarbartolo di Sciara (1987). Notarbartolo di Sciara et al. (2016) provides a detailed redescription of M. kuhlii, with only some limited remarks on how it differs from M. ereegoodootenke. The key to species provided in that paper lists the following characters as separating these two species: ventral pectoral fin coloration, length of cephalic lobes, dorsal fin tip coloration and branchial filter plates. It is puzzling though that the genetic results presented in Henderson et al. (2016), which failed to detect any differences in structure of the NADH2 marker between these two species, are not referred to in this paper despite being from the same region. Thus, it seems that Notarbartolo di Sciara et al. (2016) did not consider available information regarding the lack of genetic differentiation between M. kuhlii and M. ereegoodootenke from Oman.
Remarks: Mobula kuhlii and M. eregoodootenkee have previously been considered distinct, based primarily on the length of the cephalic lobes. It is proposed herein that the relative length of the cephalic lobes is a variable, intraspecific character, based on individuals possessing very long cephalic lobes being sampled together with those with very short lobes at the same location, for example, off Oman. The reported maximum size for the two species is similar (~100 vs. 119 cm DW). One character that has caused some confusion in these species is the presence or absence of a white tip or extent of a whitish hue on the dorsal fin. Mobula eregoodootenkee is reported to have either a plain dorsal fin (or with a whitish hue), while M. kuhlii has been reported as both with and without a white spot on the dorsal fin apex (Notarbartolo di Sciara et al. 2016). However, this character is variable. For example, two pregnant female M. kuhlii reported from Indonesia had plain greyish dorsal fins, but they each contained a single, late term embryo that had a distinct white spot on the apex of the dorsal fin. Notarbartolo di Sciara (1987) found that M. kuhlii and M. eregoodootenke shared the following characters which are diagnostic for mobulid species: no caudal spine, base of tail quadrangular in cross-section, spiracles subcircular and located ventral to plane of pectoral fins, tooth bands about three-quarters of mouth width, teeth wider than long and anterior margin of pectoral fin straight. Although some differences were noted in the tooth morphology, this was based on an adult male M. eregoodootenkee and juvenile males of M. kuhlii.
In a number of specimens of devilrays identified as M. eregoodootenkee, the pectoral fins have a blackish anterior margin with a broader blackish marking on central anterior margin (Fig. 7a). In contrast, most short-headed forms of M. kuhlii that were examined lacked these, but instead the distal portion of the ventral pectoral fins was dusky (Fig. 7b), versus white in the above specimens (Fig. 7a). This character was also reported by Notarbartolo di Sciara et al. (2016) as one of the key characters to distinguish between M. kuhlii and M. eregoodootenkee. Since coloration can be highly variable in a number of myliobatoid rays, for example, melanistic forms of M. birostris and M. alfredi, further investigation into the validity of this character is needed with examination of a larger number of specimens. As with the length of the cephalic lobes, we consider this difference to be related to intraspecific variation and not an interspecific character. Paig-Tran et al. (2013) found that the branchial filter plates differed between M. eregoodootenkee and M. kuhlii with the terminal lobe being far more elongate in M. eregoodootenkee. This character was not confirmed with the specimens examined in this study. It should be noted that only a single specimen was available for both species in the Paig-Tran et al. (2013) study, thus intraspecific variation could not be taken into account. Notarbartolo di Sciara et al. (2016) stated that the terminal lobes on the filter plates of M. kuhlii were leaf-shaped to spade-shaped, but it was not stated how many specimens were dissected to view this character. No additional information on the filter plates of M. eregoodootenkee was provided. Therefore, as with the other characters that have previously been used to separate these species, it is poorly understood how these characters vary within species.
Ventral coloration of: (A) long-head form of Mobula kuhlii (originally identified as Mobula eregoodootenkee), adult male from Muttrah in Oman; (B) short-head form of Mobula kuhlii, female from Muttrah in Oman.
The genetic analyses undertaken in this study show that specimens identified morphologically as M. kuhlii and M. eregoodootenkee are consistently very closely related based on analyses of both mitochondrial genomes and nuclear exon data (Figs 1, 2). The observed divergence between these taxa was within the range of intraspecific divergences observed for other mobulid lineages based on our expanded taxon sampling of mitochondrial NADH2 data (Supporting Information Fig. S1), an order of magnitude lower based on the mitochondrial genome data and among the lowest pairwise divergences observed for the nuclear exon data. Strengthening this argument is the presence of both long-head and short-head forms from Oman, which are indistinguishable based on sequencing of the mitochondrial NADH2 locus (Supporting Information Fig. S1; Henderson et al., 2016). In this paper, long-head forms are referred to as M. eregoodootenkee (GN9431, GN9437 and GN9438) and short-head forms are referred to as M. kuhlii (GN9426-30, GN9432, GN9678-80, GN9726, GN9729, GN9737-38 and GN9747). Thus, although there is some morphological evidence to support M. eregoodootenkee distinct from M. kuhlii, the combined morphological and detailed molecular data presented herein lead us to conclude that M. eregoodootenke is a junior synonym of M. kuhlii.
Mobula mobular (Bonnaterre, 1788)
Squalus edentulus Brünnich, 1768: 6 (Marseille, France)
Raia mobular Bonnaterre, 1788: 5 (Marseille, France)
Raja vespertilio Walbaum, 1792: 535 (Azores, northeastern Atlantic)
Aodon cornu Lacepède, 1798 (unneeded new name for S. edentulus Brünnich, 1768)
Raia aurita Suckow, 1799: 78
Raja fabroniana Lacepède, 1800: 104, 111, pl. 5 (figs 1, 2) (Livourne, Italy; holotype MZUF probably lost)
Raja cephaloptera Bloch & Schneider, 1801: 365 (no locality; holotype ZMB 13407, skull)
Raja giorna Lacepède, 1803: 662, 666, pl. 20 (fig. 3) (Bay of Nice, France)
Raja diabolus Shaw, 1804: 291 (Mediterranean, Atlantic and Indian seas)
Mobula auriculata Rafinesque, 1810: 48, 61
Apterurus fabroni Rafinesque, 1810: 48 (unjustified emendation of Raja fabroniana Lacepède, 1800)
Cephalopterus massena Risso, 1810: 15 (Nice, France)
?Raia cornuta Lesueur, 1824: 120 (based on Azores records, not from US Atlantic coast)
Cephaloptera japanica Müller & Henle, 1841: 185 (Nagasaki, Japan; lectotype RMNH D 2440; paralectotype RMNH, lost)
Cephaloptera edentula Griffini, 1903: 132, fig. 73 (Italian seas)
Mobula rancureli Cadenat, 1959: 1331, figs 1–10 (Ivory Coast; holotype MNHN 1965-0146)
Common name: Giant devilray.
Nomenclatural discussion: In order to understand the complicated taxonomic history of M. mobular, it is necessary to go back to the original record upon which several authors based their descriptions. Duhamel du Monceau (1780) provided illustrations and a description of a specimen caught in 1723 near Marseille in southern France (Mediterranean Sea). The description includes some basic morphometrics, including length from head to tail (6 ft. = ~183 cm), mouth width (15 in. = 38 cm), each wing (6 ft. = ~183 cm) and tail (4.5 ft. = ~137 cm). The dorsal and ventral illustrations provided by Duhamel du Monceau show a subterminal mouth clearly indicating a Mobula, but strangely the ventral surface depicts six gill slits on each side. In Duhamel du Monceau’s account, reference is made to the Azores where it is referred to as Raie cornue (= horned ray) and the Caribbean where it is referred to as Mobular, and some that refer to it as Squatina. In the addition section of Duhamel du Monceau (1780, 330), reference is made to Gentil’s (1779) records of Diable de mer from the Indian Ocean which he alludes to being the same as his species. Gentil’s illustration of Diable de mer, although somewhat cartoonish, agrees with the overall shape of a mobulid, but not enough key features are apparent to determine the species. Duhamel du Monceau’s Raie cornue is not considered an available name as it was used in the vernacular in reference to the horned rays of the Azores.
Raja mobular was proposed by Bonnaterre (1788) for Duhamel du Monceau’s (1780) Marseille record, with the measurements provided taken from that publication. In the same year, Schneider provided a detailed account of Duhamel du Monceau’s description of the Marseille specimen, the Azores Raie cornue, and noting Gentil’s record of Diable de Mer from the Indian Ocean. In this publication, Schneider (1788: 82, 83) proposes the name Raia vespertilio for this species in allusion to its bat-like appearance. Walbaum (1792) listed Raja vespertilio as a questionable species.
Shaw (1804) described R. diabolus based on literature sources. The primary literature source was the account of Duhamel du Monceau (1780), based on the same specimen used for the R. mobular description by Bonnaterre (1788). The specimen was stated as being 10.5 ft. long (~320 cm) which matches the measurements provided for R. mobular, that is, tail length 4.5 ft. and body length 6 ft. Shaw included Russell’s ‘Eereegoodee Tenkoo’ in the synonymy and stated that it occurs in the Mediterranean, Atlantic and Indian seas, but mainly observed around the Azores. The distribution provided is likely based on Russell’s Indian species (Indian seas), the Marseilles specimen described by Duhamel du Monceau (Mediterranean) and the Azores records (Atlantic). Based on the Mediterranean location and the large size of the Marseilles specimen, Notarbartolo di Sciara (1987) included it in the synonymy of M. mobular (Bonnaterre, 1788). Since the descriptive characters used in this description are based on the same specimen used to describe M. mobular, R. diabolus must be considered a junior synonym of this species. Interestingly, Shaw’s description states that the tail is unarmed, whereas M. mobular possesses a distinct caudal spine. Another uncertainty is the coloration which is stated by Shaw as being cinereous brown above, whereas M. mobular is typically bluish black above. It is possible that the caudal spine was removed or missing from the Marseilles specimen or simply that Duhamel du Monceau did not include that feature in his description.
Klunzinger (1871) described Dicerobatis monstrum based on a 54 cm embryo that came from a 2 m female specimen stranded at Al-Qusair, Egypt, in the Red Sea. The holotype was listed as not found and probably lost by Fricke (1992). Dor (1984) considered this species a junior synonym of M. diabolus. The description includes reference to the spiracles being located behind the eyes on the back (i.e. not beneath the pectoral-fin origin) and the dorsal colour being blue black. These characters are adequate to confirm the identity of this species as M. mobular.
In 1841, Müller & Henle described a new species, Cephaloptera japanica, based on two specimens collected off Nagasaki in Japan. The brief description provides mostly generic-level features, but examination of the dried lectotype (Fig. 8) revealed the following key diagnostic features: white-tipped dorsal fin, serrated caudal spine behind dorsal fin, spiracles located above pectoral-fin origin, pectoral fins not strongly falcate and their anterior margins nearly straight (not concave). Mobula japanica is currently recognized as a wide-ranging, almost circumtropical species which is reported to attain at least 310 cm DW. In a revision of the genus, Notarbartolo di Sciara’s (1987) states that M. japanica does not differ substantially in any characters from M. mobular from the Mediterranean. The only character found to differ was the morphology of the rete mirabile cranica and possibly in tooth morphology, but this was based on limited data. Although retaining them as separate species, Notarbartolo di Sciara states that this is only due to lack of direct examination of a sufficient number specimens. This study provides new information to support the synonymization of these two species, with precedence given to M. mobular (Bonnaterre, 1788) and Mobula japanica a junior synonym.
Dorsal view of the dried lectotype of Mobula japanica (RMNH D2440, juvenile male ~65 cm DW).
Cadenat (1959) described Mobula rancureli based on a single 2.4 m DW individual from off the Ivory Coast. Although considered a synonym of M. japanica by Notarbartolo di Sciara (1987), McEachran & Séret (1990) considered it to be a valid species. Cadenat (1959) distinguished this species from M. mobular based on the number of teeth and denticle morphology. Given this is only based on one specimen and these characters can vary greatly intraspecifically, this species should be considered a synonym of M. mobular.
A number of other names have been considered junior synonyms of M. mobular:
Squalus edentulus was described by Brünnich (1768). The brief Latin description includes ‘Squalus capite lato, plano, maxillis osseis edentulis, superiore longiore, lateribus capitis prominentibus’ which roughly translates to ‘Squalus with a wide head, flat, toothless bony jaws, the long sides of the head are prominent’. It was based on a specimen from Marseille which had head width equal to 3 ‘span’ (~68 cm based on a span equalling 9 in.), thus was referring to a large species. There is also reference to the upper jaw being file-like (rough surface) which possibly indicates that teeth are present which would rule out M. alfredi and M. birostris, which lack teeth in the upper jaw. Based on the location and size, this species is probably a synonym for M. mobular. Despite being an older name, S. edentulus has not been used as a valid name for this species and prevailing usage should be given to M. mobular (Bonnaterre, 1788).
Aodon cornu Lacepède, 1798 is an unneeded new name for S. edentulus Brünnich, 1768.
Raia aurita Suckow, 1799 appears to be based on Duhamel du Monceau’s specimen, as the latter record is included in the synonymy and the briefly described specimen is of the same proportions as the Marseille specimen (i.e. each wing 6 ft. wide). Bonnaterre’s Mobular is also listed in the synonym together with R. vespertilio. This name should be considered synonymous with M. mobular as it is evident that it is based on the same literature sources as previous names and thus is an unneeded replacement name for M. mobular Bonnaterre, 1788.
Raja cephaloptera Bloch & Schneider, 1801, also refers to the Duhamel du Monceau specimen from Marseille and provides a type (only the forepart of the skull) which is from Schneider in Leipzig also. This species was stated to occur mostly in the Pacific.
Raja fabroniana was described and illustrated by Lacepède (1800) from off Livorno, Italy. The illustration clearly depicts a large Mobula with a subterminal mouth and the large size (~4 m DW) and Mediterranean location confirm it as a junior synonym of M. mobular.
Raja giorna was described by Lacepède (1803) from the Bay of Nice in southern France. The illustration provided for this species includes a serrated caudal spine, thus confirming it as a junior synonym of M. mobular.
Cephalopterus massena was described by Risso (1810) from off Nice in southern France based on a large specimen, but with no types designated and no illustration provided. The large size and Mediterranean location strongly suggest this to be a junior synonym of M. mobular.
Apterurus fabroni Rafinesque, 1810 is an unjustified emendation of Raja fabroniana Lacepède, 1800.
Mobula auriculata Rafinesque, 1810 is based on Raja mobula of Lacepède according to Eschmeyer (2015).
Raia cornuta is considered a new name combination in Lesueur (1824), in his account of Cephaloptera giorna from off the Atlantic US coast, in reference to the Azores species. It does not appear that Lesueur intended to provide a new name combination and given it is based on the Azores species, it is possibly a synonym of M. mobular. However, it should be noted that the species referred to by Lesueur as C. giorna from the US coast is considered a synonym of Mobula sp. cf. birostris (Marshall et al., 2009). It is most likely that Lesueur was referring to Duhamel du Monceau’s Raie cornue, upon which a number of the synonyms of M. mobular were based.
Cephalopterus edentula Griffini, 1903, was a new name combination for Brünnich’s Squalus edentulus, thus is also a junior synonym of M. mobular.
Remarks: Mobula mobular and Mobula japanica were previously considered distinct species. In Notarbartolo di Sciara’s detailed 1987 revision of the genus, he stated that the two species are very similar and possibly conspecific, but retained them as separate species pending more information. Due to the large size of these species, obtaining accurate material for comparison is relatively difficult. However, recently acquired molecular information has provided critical new information. These two taxa are very closely related based on the analyses of both mitochondrial genomes and nuclear exons (p-distance = 0.002 and 0.005 for mitogenome and nuclear datasets, respectively, Figs 1, 2). Moreover, the expanded sampling of NADH2 data for these taxa revealed genetic distances that were consistently within the range of intraspecific divergences that were observed for taxa with uncontested specific status. Mobula mobular is considered to be a larger species with a maximum width of 520 cm, but mostly smaller (Ebert & Stehmann, 2013) while M. japanica is reported to attain only 310 cm DW. However, this variation in maximum size is not an adequate specific character and likely reflects the variability in maximum size in large members of this family, as evidenced by M. birostris (Walbaum, 1792) which has been reported to be as large as 910 cm DW, but is rarely found over 600 cm DW (Marshall et al., 2009).
The only characters provided by Notarbartolo di Sciara (1987) to distinguish these two species were the morphology of the rete mirabile cranica and the teeth, but these were based only on a single specimen. Teeth can vary greatly in morphology within a species depending on size and sex. The teeth of M. japanica examined by Notarbartolo di Sciara (1987) were from two adult specimens, while examination of the teeth of M. mobular was based on a single juvenile specimen. Based on the lack of any substantial distinguishing features to separate these two species and the genetic results available, these two species are herein considered conspecific, with M. mobular (Bonnaterre, 1788) given precedence.
Other available mobulid names
The following available binomial names for the Mobulidae cannot be accurately assigned to recognized species based on the limited descriptive data available in the original descriptions:
Raja monstrosa Walbaum, 1792 was listed as a questionable species by Walbaum (1792) without location. The characters provided do not allow its identity to be determined and thus should be considered nomen dubium.
Raja banksiana Lacepède, 1800 was described from an illustration of a specimen observed in the East Indies (Indonesia?). The illustration is somewhat cartoonish, with the cephalic lobes both with long filaments, eyes located dorsally, no dorsal fin and elongate markings on the dorsal surface. But Lacepède noted that the illustration of this species was drawn from a specimen swimming, and thus the cephalic filaments are likely the result of distortion through the sea surface. As a result, the identity of the mobulid species drawn cannot be accurately determined and should be considered nomen dubium.
Raja barbata was briefly described by Bloch & Schneider (1801) from the African sea, but the characters provided do not allow for accurate identification of the species and thus should be considered nomen dubium.
Ceratoptera lesueurii was designated in Swainson (1839), as a footnote in a brief description of the genus Ceratoptera Müller & Henle, 1837. The illustration of the head clearly depicts a Manta species, but it is not possible to determine which species and thus should be considered nomen dubium.
DISCUSSION
The analyses of the mitochondrial genome and nuclear exon data produced topologically similar results. The overall cladistic structure is also topologically similar to previous inferences of mobulid phylogeny based on morphology (Adnet et al., 2012; Paig-Tran et al., 2013) and molecular data (Aschliman, 2011; Naylor et al., 2012b; Poortvliet et al., 2015), with the exception of the novel placement of M. tarapacana (discussed below).
Based on our analyses, Mobula alfredi and M. birostris, previously placed in the genus Manta, are nested within the other Mobula species and sister to M. mobular. That the genus Mobula is rendered paraphyletic by the inclusion of Manta has been suggested many times previously in the literature, based on both morphological (Herman et al., 2000; Adnet et al., 2012; Paig-Tran et al., 2013) and molecular (Aschliman, 2011, 2014; Naylor et al., 2012b; Poortvliet et al., 2015) inference. In contrast to the comparatively sparse genomic samplings of nuclear data that have been considered previously for this group (Aschliman 2014; Poortvliet et al., 2015), our nuclear phylogeny is based on hundreds of independent nuclear orthologs and is fully resolved. Importantly, we are thus able to demonstrate, for the first time, that this relationship holds true based on inferences from the nuclear genome and is therefore unlikely to be the consequence of inadequate genomic sampling, mitochondrial introgression or lineage sorting issues. The inferred topology suggests that the terminal mouth that is present in M. birostris and M. alfredi is a derived character within Mobula.
It should be noted that, of all our pairwise species comparisons, the comparison between M. birostris and M. alfredi was consistently among the most shallow of observed divergences. The observed genetic distance between this pair was, at times, lower than that observed between other species pairs which we have synonomized here. However, unlike these other cases, the taxonomy of M. birostris and M. alfredi has recently been revised based on morphological and meristic data (excepting the genus name change that we detail here). Moreover, a phylogeographic study has provided evidence that these species diverged relatively recently and have experienced post-divergence gene flow. It is thus already documented that these species are indistinguishable based on mitochondrial DNA (Kashiwagi et al., 2012), consistent with our own results. Although morphology was used to confirm the species identification of the specimens that we subjected to our nuclear gene capture protocols, and care was taken to select individuals from regions where the two species do not co-occur, it is likely that recent divergence possibly combined with the inclusion of introgressed individuals in our analysis is driving the shallow divergence we observe between this species pair. Obviously, elucidating the pattern of hybridization between these species is beyond the scope of this paper and is being addressed elsewhere. This result however does not impact our conclusion that Manta is an invalid generic name.
Mobula eregoodootenkee has been synonomized with M. kuhlii and we suggest that the character that has been considered diagnostic in these species – the relative length of the cephalic lobes – is in fact a variable trait in this single species. It is acknowledged that it is possible that these species are discrete and that the close relationship observed in our genetic data is the consequence of hybridization between them, as is observed between M. birostris and M. alfredi. However, we make some important distinctions between this case and the former. Firstly, M. kuhlii is considered an uncommon species that is designated data deficient by the International Union for the Conservation of Nature Red List of Threatened Species. In particular, it is noted that the range of this species is poorly documented. However, the records that do exist show M. kuhlii to be partially overlapping the distribution of M. eregoodootenkee. Secondly, a review of the nomenclature indicates that both M. eregoodootenkee and M. kuhlii have a convoluted history of being mistaken for each other and other Mobula species, suggesting ambiguity in distinguishing them. While cephalic lobe length has been used to distinguish these species, insufficient comparative material has prevented a thorough examination of variation in this trait. Likewise, suggestions that other traits such as branchial filter plate morphology may also differ between these species, have been based on limited samples (Paig-Tran et al., 2013) and thus cannot be considered diagnostic. Finally, molecular data have not previously been taken into consideration. Poortvliet et al. (2015) did not discuss the taxonomic implication of the close relationship they observed between M. eregoodootenkee and M. kuhlii based on their mitochondrial inference. Henderson et al. (2016) did note that M. eregoodootenkee and M. kuhlii sampled from southeast Arabia were genetically indistinguishable based on sequencing of the mitochondrial NADH2 gene, which is demonstrated further by our own analyses. We now demonstrate that these taxa are virtually indistinguishable based on sequence data from a large number of nuclear and mitochondrial genes. In the case of M. birostris and M. alfredi, the taxonomy was well resolved (Marshall et al., 2009) and a subsequent population-level molecular study revealed them to be introgressed for mitochondrial DNA (Kashiwagi et al., 2012). In this case, we are dealing with complex nomenclatural history that is based on potentially ambiguous morphological characters and now a large amount of molecular data suggest that these taxa are conspecific. While it is possible that a future large, population level, comparative study of both species based on molecular and morphological characters may reveal that they are distinct, potentially hybridizing species, the weight of the current evidence does not suggest this. We argue that obtaining access to such sample sizes is unlikely for these species and so their taxonomy should reflect the best available evidence, which is to synonymize these species, as we have done.
Several authors have noted previously that M. mobular and M. japanica may be conspecific based on morphological data (Notarbartolo di Sciara, 1987; Paig-Tran et al., 2013). Poortvliet et al., (2015) also noted a lack of significant differentiation between this species pair based on their sequencing of mitochondrial DNA, but elected to retain the current taxonomy in the interest of stability. In this paper, we have made the important addition of investigating this relationship using a large panel of nuclear molecular makers and have again come to the conclusion that these taxa appear to be conspecific. While we acknowledge that our sampling of M. mobular is limited, again we must argue that this is a pervasive problem for these animals that is unlikely to improve in the near future. In light of this, the taxonomy should reflect the best available evidence of the time. In the case of this species pair, a complex nomenclatural history is now combined with inferences from both morphology and molecular data to suggest conspecificity and we have thus revised the taxonomy to reflect this. It should also be noted here that the taxonomic baseline that exists for the Mobulidae has impacted our understanding of the group. That is, if M. mobular and M. japanica had been synonomized over a century ago, there would by no means be enough evidence available to separate the species today based on the information at hand. But since they have been considered as distinct species, far more evidence is required to clarify their validity. For a group such as mobulids, accessing large numbers of samples is very unlikely, thus it is important to have resolution based on the best available information. In this case, the evidence towards keeping these species separate is very weak, while the evidence to synonymize is quite strong.
Previous studies of the relationship between M. rochebrunei and M. hypostoma have been based on limited sampling, as is the case in our present genetic analysis. However, we feel that our analysis makes a substantial contribution to our understanding of this relationship that justifies the taxonomic changes that were made. Poortvliet et al. (2015) compared these taxa on the basis of a single mitochondrial gene. Here, we are able to extend genomic sampling to include inferences based on the protein coding components of whole mitochondrial genomes, and again show that these taxa are indistinguishable. While more extensive sampling of both taxa would be ideal to investigate this relationship further, this is unrealistic in the case of the extremely rare M. rochebrunei. We also acknowledge that we were unable to obtain nuclear data for comparison in this instance, but such are the limitations of molecular technologies as applied to archival material, at this point in time. Even in light of our own limitations in sampling for this comparison, the holotype of M. rochebrunei has now been confirmed to be identical to M. hypostoma at the mitochondrial genome level. Given that it is highly unlikely that anyone will obtain a substantial number of samples from either taxon, and we are faced with the complexities of obtaining sufficient high-quality nuclear data from the holotype specimen of M. rochebrunei, we believe that this is compelling evidence that these samples are, in fact, conspecific with the newer M. rochebrunei becoming a junior synonym of M. hypostoma. If future additional sampling shows that there is indeed a distinct small species in the Eastern Atlantic, then this would constitute an as yet undescribed species. In the absence of additional material (samples and nuclear data), we feel that applying the precautionary principle would constitute acknowledging this lineage as a single species with the possibility of population structure across the Atlantic Ocean.
Morphological characters that map to the molecular phylogenetic inferences that we present are summarized in Table 1. Several synapomorphic traits link the M. kuhlii–M. thurstoni and M. hypostoma–M. munkiana clades relative to the M. alfredi–M. birostris–M. mobular–M. tarapacana clades. These are position of the spiracles relative to plane of the pectoral disc (ventral vs. dorsal), presence of denticles on dorsal body (sparse vs. dense) and maximum size (< 2 m vs. > 3 m). The trait which distinguishes M. kuhlii and M. thurstoni from M. hypostoma and M. munkiana is the width of the lower tooth band, that is, ≤ 60% of mouth width versus > 75% of mouth width.
Key morphological characters amongst the species of Mobula
| . | Spiracle position relative to plane of disc . | Denticles on dorsal body . | Maximum disc width . | Width of lower tooth band (as % of mouth width) . | Caudal sting . | Presence of tooth bands . | Mouth position on head . | Branchial filter plates . |
|---|---|---|---|---|---|---|---|---|
| M. hypostoma | ventral | sparse | 1.2 m | ~47% | absent | both jaws | subterminal | separate |
| M. munkiana | ventral | sparse | 1.1 m | ~60% | absent | both jaws | subterminal | separate |
| M. kuhlii | ventral | sparse | 1.35 m | ~78% | absent | both jaws | subterminal | separate |
| M. thurstoni | ventral | sparse | 1.89 m | ~76% | absent | both jaws | subterminal | separate |
| M. alfredi | dorsal | dense | 5.5 m | up to 77% | absent | lower jaw only | terminal | separate? |
| M. birostris | dorsal | dense | 7 m | up to 70% | encased in calcified mass | lower jaw only | terminal | separate? |
| M. mobular | dorsal | dense | 5.2 m | ~76% | present | both jaws | subterminal | separate |
| M. tarapacana | dorsal | dense | 3.7 m | ~70% | absent | both jaws | subterminal | fused together |
| . | Spiracle position relative to plane of disc . | Denticles on dorsal body . | Maximum disc width . | Width of lower tooth band (as % of mouth width) . | Caudal sting . | Presence of tooth bands . | Mouth position on head . | Branchial filter plates . |
|---|---|---|---|---|---|---|---|---|
| M. hypostoma | ventral | sparse | 1.2 m | ~47% | absent | both jaws | subterminal | separate |
| M. munkiana | ventral | sparse | 1.1 m | ~60% | absent | both jaws | subterminal | separate |
| M. kuhlii | ventral | sparse | 1.35 m | ~78% | absent | both jaws | subterminal | separate |
| M. thurstoni | ventral | sparse | 1.89 m | ~76% | absent | both jaws | subterminal | separate |
| M. alfredi | dorsal | dense | 5.5 m | up to 77% | absent | lower jaw only | terminal | separate? |
| M. birostris | dorsal | dense | 7 m | up to 70% | encased in calcified mass | lower jaw only | terminal | separate? |
| M. mobular | dorsal | dense | 5.2 m | ~76% | present | both jaws | subterminal | separate |
| M. tarapacana | dorsal | dense | 3.7 m | ~70% | absent | both jaws | subterminal | fused together |
Key morphological characters amongst the species of Mobula
| . | Spiracle position relative to plane of disc . | Denticles on dorsal body . | Maximum disc width . | Width of lower tooth band (as % of mouth width) . | Caudal sting . | Presence of tooth bands . | Mouth position on head . | Branchial filter plates . |
|---|---|---|---|---|---|---|---|---|
| M. hypostoma | ventral | sparse | 1.2 m | ~47% | absent | both jaws | subterminal | separate |
| M. munkiana | ventral | sparse | 1.1 m | ~60% | absent | both jaws | subterminal | separate |
| M. kuhlii | ventral | sparse | 1.35 m | ~78% | absent | both jaws | subterminal | separate |
| M. thurstoni | ventral | sparse | 1.89 m | ~76% | absent | both jaws | subterminal | separate |
| M. alfredi | dorsal | dense | 5.5 m | up to 77% | absent | lower jaw only | terminal | separate? |
| M. birostris | dorsal | dense | 7 m | up to 70% | encased in calcified mass | lower jaw only | terminal | separate? |
| M. mobular | dorsal | dense | 5.2 m | ~76% | present | both jaws | subterminal | separate |
| M. tarapacana | dorsal | dense | 3.7 m | ~70% | absent | both jaws | subterminal | fused together |
| . | Spiracle position relative to plane of disc . | Denticles on dorsal body . | Maximum disc width . | Width of lower tooth band (as % of mouth width) . | Caudal sting . | Presence of tooth bands . | Mouth position on head . | Branchial filter plates . |
|---|---|---|---|---|---|---|---|---|
| M. hypostoma | ventral | sparse | 1.2 m | ~47% | absent | both jaws | subterminal | separate |
| M. munkiana | ventral | sparse | 1.1 m | ~60% | absent | both jaws | subterminal | separate |
| M. kuhlii | ventral | sparse | 1.35 m | ~78% | absent | both jaws | subterminal | separate |
| M. thurstoni | ventral | sparse | 1.89 m | ~76% | absent | both jaws | subterminal | separate |
| M. alfredi | dorsal | dense | 5.5 m | up to 77% | absent | lower jaw only | terminal | separate? |
| M. birostris | dorsal | dense | 7 m | up to 70% | encased in calcified mass | lower jaw only | terminal | separate? |
| M. mobular | dorsal | dense | 5.2 m | ~76% | present | both jaws | subterminal | separate |
| M. tarapacana | dorsal | dense | 3.7 m | ~70% | absent | both jaws | subterminal | fused together |
Several characters that were previously considered important for understanding relationships among mobulid species, such as the absence of tooth bands in the lower jaw, position of the mouth on head and presence of a caudal sting, are herein considered to be plesiomorphic traits. They are thus not useful characters for determining relationships amongst species within the Mobulidae.
The variable position of M. tarapacana inferred from the analyses of the mitochondrial versus nuclear datasets is particularly interesting. Mobula tarapacana shares some traits with M. alfredi, M. birostris and M. mobular, such as the dense covering of denticles on dorsal body, dorsal position of spiracles relative to plane of disc and large maximum size (Table 1). These traits thus appear to support the inferences based on the mitochondrial genome data (Fig. 1) that place M. tarapacana sister to M. alfredi, M. birostris and M. mobular. Inferences based on the nuclear exon data place M. tarapacana basal to all other mobulids with strong bootstrap support (Fig. 2). On the one hand, mitochondrial DNA has a smaller effective population size than the nuclear genome and is thus expected to sort more rapidly. That being said, mitochondrial DNA is inherited as a single linked unit; thus, even analyses of whole mitochondrial genome data can only be considered a single locus and the possibility that this inference reflects gene tree–species tree discordance cannot be ruled out. It should be noted that the placement of M. tarapacana inferred from the mitochondrial DNA data has lower bootstrap support than other relationships in the tree (Fig. 1). Inferences resulting from both concatenated ML and coalescent-based analyses of the complete and clock-filtered datasets, which included data from 1082 and 614 nuclear exons, respectively, consistently resolved M. tarapacana as basal to all other mobulids with strong bootstrap support, thus providing compelling evidence for this relationship. Mobula tarapacana does possess several traits that make it a relatively unique species within the Mobulidae. Most importantly, the branchial filter plates are fused together in M. tarapacana, but are separate in other mobulid species. This species also possesses unique tooth morphology and both of these characters have been used to suggest that M. tarapacana is basal to other mobulids (Herman, 2000; Adnet et al. 2012; Paig Tran et al., 2013). Other unique traits include a strong ridge on the midline of the body, very falcate pectoral fins, and a short tail that is rigid and rod-like. As mentioned previously, very few whole mobulid specimens that are in sufficiently good condition to enable more detailed morphological comparisons can be found in museum collections. This, together with the mito-nuclear discordance in the placement of M. tarapacana presented in this study, suggests that more detailed anatomical studies of M. tarapacana in relation to the other mobulids are required. For example, preliminary investigation into the structure of the vertebrae beneath the first dorsal fin of several mobulid species in Indonesia revealed that M. tarapacana possessed extremely small vertebrae encased in a very large haemal arch compared to M. mobular, M. kuhlii and M. thurstoni. How this character and other skeletal characters differ between the other mobulid species may be important to better understand these relationships.
The extremely close relationship between M. munkiana and M. hypostoma that was revealed by analyses of the nuclear exon data was surprising. These analyses suggested that these two species were more closely related than are M. eregoodootenkee and M. kuhlii, M. japanica and M. mobular, and M. alfredi and M. birostris. The main character used to separate M. munkiana from M. hypostoma is the width of the lower tooth band (60% vs. 47%; Table 1). Thus, further taxonomic research focused on these two species is required to ascertain whether they are truly separate species, or whether the differences reflect population variation within a single more widely ranging species.
Key to the genus Mobula
1. Mouth terminal on head ………………………...…. 2
– Mouth subterminal on ventral surface of head ……………………………………………………….….. 3
2. Calcified mass containing an embedded spine present behind dorsal fin; anterior margin of white shoulder patches running parallel with front of head; a large black, semicircular spot emanating from both of the fifth gill slits ……. Mobula birostris
– No calcified mass or spine present behind dorsal fin; anterior margin of white shoulder patches curving posteriorly and not parallel with front of head; a small black, semicircular spot emanating from both of the fifth gill slits ………….....…… Mobula alfredi
3. Spiracles long, slit-like and dorsal to the plane of the pectoral fins; large species reaching well over 200 cm DW …………………………………….……… 4
– Spiracles small, subcircular and ventral to the plane of the pectoral fins; small species, not reaching 200 cm DW and mostly less than 130 cm DW ……………………………………………….........……. 5
4. Pectoral fins strongly falcate; a strong bony ridge present along dorsal midline; no caudal spine present; disc greyish, dorsal fin plain; branchial filter plates on gill arches fused along their lateral margins ……………………....……… Mobula tarapacana
– Pectoral fins moderately falcate; bony ridge on dorsal midline weak; a serrated caudal spine usually present behind dorsal fin; disc blackish, dorsal fin with prominent white tip; branchial filter plates on gill arches separate (not fused) along their lateral margins …………….........……........ Mobula mobular
5. Tooth bands in each jaw less than 65% of mouth width …………………………….…………………….. 6
– Tooth bands in each jaw more than 65% of mouth width …………………….......………………………… 7
6. Lower tooth band length about 60% of mouth width ……......……………………………. Mobula munkiana
– Lower tooth band length about 47% of mouth width ………………….......……………... Mobula hypostoma
7. Anterior margin of pectoral fins with a double curvature (hence common name of Bentfin Devilray); base of tail depressed; dorsal coloration bluish black; dorsal fin with a prominent white tip; attains about 190 cm DW …………...................... Mobula thurstoni
– Anterior margin of pectoral fins straight or slightly convex; base of tail quadrangular; dorsal coloration greyish brown; dorsal fin with or without a white tip; attains about 135 cm DW ……………………………………......... Mobula kuhlii
Material examined
Mobula alfredi (Krefft, 1868)
Not retained, tissue sampled: Tissue accessions GN15457, GN15458, GN15459 and GN15460, South Africa.
All specimens were nominally identified as M. alfredi.
Mobula birostris (Walbaum, 1792)
CSIRO H 6446-02 (one clasper only), adult male 461.4 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 18 August 2005; CSIRO H 7791-01 (dorsal fin only), CSIRO H 7791-02 (dorsal fin only), Tanjung Luar landing site, Lombok, Indonesia, 5 October 2009; field code BRU-043, Philippines, 20 April 2007.
Photographed (not retained): male embryo 60.9 cm DW, Pelabuhanratu landing site, West Java, Indonesia; female 493 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 25 March 2002; female 412.6 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 25 April 2004; female 337.6 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 26 April 2004; female 377.4 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 13 July 2004; adult male 340.6 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 9 March 2005; male 289.1 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 11 March 2005; adult male 396 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 13 July 2005; male 270 cm DW (tissue accession GN6791), Cilacap landing site, Central Java, Indonesia, 19 October 2008.
Not retained, tissue sampled: Tissue accessions GN10559, GN10560, GN10561, GN10568, Japan, 26 September 2012.
All specimens were nominally identified as M. birostris.
Mobula hypostoma (Bancroft, 1831)
BMNH 1897.7.1.40 (holotype of Ceratobatis robertsii), juvenile female 92 cm DW, Jamaica.
MNHN A9967 (dry, holotype of C. rochebrunei; tissue accession GN13192), adult male 108.5 cm DW, Senegal.
Photographed (not retained): Field code MS05-390 (tissue accession GN5813), female 73 cm DW, field code MS05-391 (tissue accession GN5814), female 74 cm DW, St Joes Bay, Gulf of Mexico, Florida, USA, 3 October 2006.
Mobula kuhlii (MÜller & Henle, 1841)
BMNH 1870.8.31.68-69 (syntypes of Dicerobatis draco), 2 male embryos, 36.8 and 34.6 cm DW, Misol Island, West Papua, Indonesia; CAS 56095 (neotype of M. eregoodootenkee), adult male 96.9 cm DW, Gulf of Siam, Cambodia, 5.5 m depth, 26–27 March 1961; CSIRO H 7776-01 (tissue accession GN 11296), embryo 31 cm DW, CSIRO H 7776-02 (tissue accession GN 11297), embryo 23.8 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 27 February 2011; MNHN 0000-1596 (lectotype), juvenile male 71.7 cm DW, 70.6 cm TL, India; MZB 15036 (tissue accession GN 11235), male 116 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 8 June 2002; NMW 77991; field code BRU-031 (tissue accession GN4337), male 132.5 cm DW, Iloilo City fishing port, Philippines, 13 February 1999.
Photographed (not retained): Field code HBO122 (tissue accession GN3019), male 114 cm DW, Tawau, Sabah, Malaysia, 28 June 2002; field code BO15, male 80.5 cm DW, Sematan, Sarawak, Malaysia, 1 June 2002; field code MM-835, female 92 cm DW, Gulf of Oman, Iran, 22 September 2010; pregnant females 135.2 and 134.2 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 27 February 2011; male 104.1 cm DW, Sabah, Malaysia; adult male 105 cm DW, Al Khor, Qatar, 24 April 2009; female ~100 cm DW, northern Australia; adult male, Muttrah, Oman, 20 September 2010.
Not retained, tissue sampled: Field code JPAG 303 (tissue accession GN4327), Philippines, 18 April 2007; tissue accession GN9426, female, tissue accession GN9427, male, tissue accession GN9428, male, tissue accession GN9429, male, tissue accession GN9430, female, tissue accession GN9431 (nominally identified M. eregoodootenkee), female, tissue accession GN9432, male, Muttrah, Oman, 1 March 2011; tissue accession GN9437 (nominally identified M. eregoodootenkee), female, Muttrah, Oman, 4 December 2010; tissue accession GN9580, Barka, Oman, 7 December 2011; tissue accession GN9678 and GN9679, Arabian Sea coast of Oman; tissue accession GN9680, Sharjah, Oman; tissue accession GN9726, female, Muttrah, Oman, 29 May 2010; tissue accession GN9729, female, Muttrah, Oman, 24 August 2011; tissue accession GN9737, male, tissue accession GN9738, male, Muttrah, Oman, 20 September 2010; tissue accession GN9747, female, Muttrah, Oman, 23 August 2011; tissue accessions GN15461 (nominally identified M. eregoodootenkee), GN15462 (nominally identified M. eregoodootenkee), GN15463 (nominally identified M. eregoodootenkee), GN15464 (nominally identified M. eregoodootenkee), GN15465 (nominally identified M. eregoodootenkee), South Africa.
Mobula mobular (Bonnaterre, 1788)
CSIRO H 6139-01 (tissue accession GN11234, nominally identified M. japanica), juvenile female 64.5 cm DW, Pelabuhanratu landing site, West Java, Indonesia, 16 August 2002; CSIRO H 6446-01 (one clasper only), adult male 273.6 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 18 August 2005; CSIRO H 6660-05 (dorsal fin and tail only; tissue accession GN11233, nominally identified M. japanica), male, Cilacap landing site, Central Java, Indonesia, 10 June 2002; CSIRO H 5885-04 (dorsal fin only), female, Tanjung Luar landing site, Lombok, Indonesia, 8 June 2002; CSIRO H 5885-05 (gill arch only), adult male, Tanjung Luar landing site, Lombok, Indonesia, 8 June 2002; CSIRO H 7775-03, 84 cm DW, 183.2 cm TL, Tanjung Luar landing site, Lombok, Indonesia, 7 October 2009; HUMZ 96485, juvenile female 108.5 cm DW, 192.9 cm TL, location unknown; HUMZ 144781, female 112 cm DW, 175 cm TL, Usujiri, Japan, August 1996; MZB 15028 (tissue accession GN11232, nominally identified M. japanica), female 107.8 cm DW, Kedonganan fish market, Bali, Indonesia, 4 June 2002; RMNH D2440 (dry; lectotype of M. japanica), juvenile male ~65 cm DW, Nagasaki, Japan.
Photographed (not retained): Field code PR3, female 92 cm DW, Pelabuhanratu landing site, West Java, Indonesia, 20 April 2001; field code BJ-391 (tissue accession GN5273, nominally identified M. japanica), female 207 cm DW, La Paz, Gulf of California, Mexico, 8 September 1993; field code BJ-409 (tissue accession GN5276, nominally identified M. japanica), female 209 cm DW, field code BJ-410 (tissue accession GN1562, nominally identified M. japanica), female 191 cm DW, La Paz, Gulf of California, Mexico, 11 September 1993; field code BJ-432 (tissue accession GN5287, nominally identified M. japanica), male 200 cm DW, La Paz, Gulf of California, Mexico, 26 September 1993; field code BJ-435 (tissue accession GN5289, nominally identified M. japanica), 202 cm DW, La Paz, Gulf of California, Mexico, 27 September 1993; field code BJ-772 (tissue accession GN1558, nominally identified M. japanica), female 221 cm DW, BJ-773 (tissue accession GN1559, nominally identified M. japanica), male 212 cm DW, Punta Arena, Gulf of California, Mexico, 22 June 1996; field code BJ-774, male 218 cm DW, field code BJ-775 (tissue accession GN5433, nominally identified M. japanica), female 219.5 cm DW, Punta Arena, Gulf of California, Mexico, 24 June 1996; field code BJ-775 (tissue accession GN5434, nominally identified M. japanica), female 211 cm DW, Punta Arena, Gulf of California, Mexico, 25 June 1996; field code BJ-WWW (tissue accession GN5452, nominally identified M. japanica), male 211 cm DW, Punta Arena, Gulf of California, Mexico, 27 June 1996; field code VN-60 (tissue accession GN7058, nominally identified M. japanica), male > 222 cm DW, VN-61 (tissue accession GN7059, nominally identified M. japanica), female > 222 cm DW, Phan Thiet, Vietnam, 15 March 2010; embryo ~50 cm DW, Pelabuhanratu landing site, West Java, Indonesia, 1 July 2001; female 211 cm DW, Cilacap landing site, Central Java, Indonesia, 3 July 2001; male 206 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 26 March 2002; female 182 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 8 June 2002; juvenile male 116 cm DW, female 202 cm DW, Cilacap landing site, Central Java, Indonesia, 10 June 2002; female 122 cm DW, Kedonganan fish market, Bali, Indonesia, 22 August 2002; female 110 cm DW, Kedonganan fish market, Bali, Indonesia, 17 April 2004; adult male 210 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 26 April 2004; female 151 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 9 March 2005; juvenile male 156.5 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 13 July 2005; female 187.2 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 27 March 2006; female 179.7 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 28 March 2006; Field code PC45, 141.1 cm DW, Pacitan fish landing site, East Java, Indonesia, 17 October 2008; Field code LM353, 112.8 cm DW, 223.6 cm TL, Tanjung Luar landing site, Lombok, Indonesia, 7 October 2009; female embryo 68.2 cm DW, Pelabuhanratu landing site, West Java, Indonesia.
Not retained, tissue sampled: Tissue accession GN8642 (nominally identified M. japonica), male, Muttrah, Oman, 23 August 2011; tissue accession GN9579 (nominally identified M. japonica), male, Muttrah, Oman, 19 July 2011; tissue accession GN9681 (nominally identified M. japonica), Ras al Khaimah, Oman; tissue accession GN9727 (nominally identified M. japonica), female, Muttrah, Oman, 6 September 2010; tissue accession GN9739 (nominally identified M. japonica), female, Muttrah, Oman, 15 June 2011; tissue accession GN10565 (nominally identified M. japonica), Japan, 26 September 2012; tissue accession GN12940 (nominally identified M. japonica), Kagoshima, Japan; tissue accession GN15654, between Mallorca and Menorca Islands, Spain, 25 August 2009.
Mobula munkiana Notarbartolo di Sciara, 1987
Photographed (not retained): Field code BJ-275 (tissue accession GN2286), male 86 cm DW, Santa Rosalia, Gulf of California, Mexico, 20 August 1993; field code BJ-294, female 100 cm DW, Santa Rosalia, Gulf of California, Mexico, 20 August 1993; field code BJ-315 (tissue accession GN5255), female 90 cm DW, field code BJ-319 (tissue accession GN5257), female 86 cm DW, Loreto, Gulf of California, Mexico, 27 August 1993; field code BJ-358, male 104 cm DW, BJ-361, male 82 cm DW, Loreto, Gulf of California, Mexico, 1 September 1993.
Not retained, tissue sampled: Field code BJ-310 (tissue accession GN5251), male 87 cm DW, BJ-311 (tissue accession GN5252), female 92 cm DW, BJ-312 (tissue accession GN5253), female 99.5 cm DW, field code BJ-320 (tissue accession GN5258), female 93 cm DW, Loreto, Gulf of California, Mexico, 27 August 1993; field code BJ-413 (tissue accession GN5278), female 100 cm DW, La Paz, Gulf of California, Mexico, 11 September 1993.
Mobula tarapacana (Philippi, 1892)
CSIRO H 6660-06 (dorsal fin and tail only; tissue accession GN11236), female, Cilacap landing site, Central Java, Indonesia, 10 June 2002; CSIRO H 5885-06 (gill arch only), adult male, Tanjung Luar landing site, Lombok, Indonesia, 8 June 2002; CSIRO H 7406-04 (dorsal fin only), 278 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 4 March 2009; MNHN 1965-0144 (holotype of Mobula coilloti), male, ~10 miles south of Iridi, Ivory Coast, western Africa, 13 March 1959.
Photographed (not retained): adult male 288 cm DW, Cilacap landing site, Central Java, Indonesia, 2 July 2001; adult male 265 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 8 June 2002; female 201 cm DW, Cilacap landing site, Central Java, Indonesia, 10 June 2002; female 288.6 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 11 July 2004; female 318.2 cm DW, female 267 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 13 July 2004; male 186.7 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 12 October 2004; male 182.8 cm DW, female 219.5 cm DW, female 272.6 cm DW, female 246.9 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 19 January 2005; female 151 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 9 March 2005; male 194.2 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 10 July 2005; female 303.4 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 26 March 2006; female 258.7 cm DW, female 235.7 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 26 October 2008; 278 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 4 March 2009.
Not retained, tissue sampled: Tissue accession GN10564, Japan, 26 September 2012.
Mobula thurstoni (Lloyd, 1908)
CSIRO H 7774-01 (juvenile male 78.7 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 14 July 2005; CSIRO H 7775-01 (head only), ~210 cm DW, CSIRO H 7775-02, ~166 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 7 October 2009; HUMZ 114050, juvenile male 86 cm DW, 119.1 cm TL, Usujiri, Japan, 24 August 1989; MZB 15007, juvenile female 82.6 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 13 July 2004; MZB 15037, female 94 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 12 July 2005; MZB 15444 (tissue accession GN11237), female 91.5 cm DW, Cilacap landing site, Central Java, Indonesia, 11 June 2002; RMNH unregistered, male embryo ~42 cm DW, no collection data; SMEC KBU 1 15397, male embryo 34.7 cm DW, Sabah, Malaysia, 15 March 1997.
Photographed (not retained): Field code BJ-330 (tissue accession GN5263), male 96 cm DW, Loreto, Gulf of California, Mexico, 29 August 1993; field code BJ-360, male 83 cm DW, Loreto, Gulf of California, Mexico, 1 September 1993; field code BJ-429 (tissue accession GN5284), female 180 cm DW, La Paz, Gulf of California, Mexico, 25 September 1993; field code BJ-434, female 149 cm DW, La Paz, Gulf of California, Mexico, 26 September 1993; field code BJ-705 (tissue accession GN1560), male 172 cm DW, BJ-706 (tissue accession GN1561), male 169 cm DW, Santa Rosalia, Gulf of California, Mexico, 13 June 1996; field codes SP-10, female 115.5 cm TL, SP-11, male 126 cm TL, Gulf of California, Mexico, 18 July 1994; male 145 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 10 September 2004; female 107.1 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 12 October 2004; male 111.5 cm DW, Kedonganan fish market, Bali, Indonesia, 7 July 2005; female 113 cm DW, Tanjung Luar landing site, Lombok, Indonesia, 12 July 2005.
Not retained, tissue sampled: Field code BJ-351 (tissue accession GN5268), male 89 cm DW, Loreto, Gulf of California, Mexico, 1 September 1993; field code BJ-378 (tissue accession GN5270), male 170 cm DW, La Paz, Gulf of California, Mexico, 6 September 1993; field code BJ-411 (tissue accession GN5277), male 171 cm DW, La Paz, Gulf of California, Mexico, 11 September 1993; field code BJ-431 (tissue accession GN5286), male 172 cm DW, La Paz, Gulf of California, Mexico, 25 September 1993; tissue accession GN9725, male, Muttrah, Oman, 3 August 2010; tissue accession GN9728, male, Muttrah, Oman, 17 August 2010.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Figure S1. Maximum Likelihood tree showing nominal identifications of mobulid specimens and the relationships among them, relative to three outgroups (Aetobatus narinari, Myliobatis aquila and Rhinoptera bonasus). The tree was derived from an alignment of mitochondrial NADH2 sequences (1044 sites) under a TVM + I + G model of molecular evolution.
Table S1. Best partitioning scheme for the protein coding components of the mitochondrial genomes.
ACKNOWLEDGEMENTS
We would like to thank the following people for their contribution to this study: Alastair Graham (CSIRO) for providing collection information; John Pogonoski (CSIRO) for providing editorial feedback on the final draft of the manuscript; Carlie Devine (CSIRO) for preparing images; Alec Moore (RSK Group) for providing images from the Persian Gulf; Simon Weigmann (University of Hamburg) for helping to track down obscure references; Alan Reeve and Tariq Al-Mamari (Sultan Qaboos University) for their help with specimen collection. Kerri Matthes and Elisabeth Rochel performed data collection for the mitochondrial NADH2 dataset. We would also like to thank the following museum staff for their hospitality during museum visits by WW and for providing collection data for material examined: Mamoru Yabe, Hisashi Imamura, Toshio Kawai, Kazuhiro Nakaya, Keisuke Ogimoto and other students (HUMZ); James Maclaine and Oliver Crimmen (BMNH); Romain Causse, Bernard Séret, Guy Duhamel, Patrice Pruvost, Zora Gabsi and Claude Ferrara (MNHN); Ronald de Ruiter and Martien van Oijen (RMNH); and Renny Hadiaty (MZB). Thanks also to Janine Caira (University of Connecticut, USA) and Kirsten Jensen (Kansas University, USA) for their elasmobranch host specimen database available at http://tapewormdb.uconn.edu. Four of us (WW, SC, LY and GN) were supported by a National Science Foundation (NSF; http://www.nsf.gov) grant (Jaws and Backbone: Chondrichthyan Phylogeny and a Spine for the Vertebrate Tree of Life; DEB-01132229). The first author was also supported by the CSIRO’s Oceans & Atmosphere Flagship and the National Research Collections Australia.