-
PDF
- Split View
-
Views
-
Cite
Cite
Kate M. Serdy, José Pablo Leone, David J. Dabbs, Rohit Bhargava, Male Breast Cancer: A Single-Institution Clinicopathologic and Immunohistochemical Study, American Journal of Clinical Pathology, Volume 147, Issue 1, January 2017, Pages 110–119, https://doi.org/10.1093/ajcp/aqw207
Close - Share Icon Share
Objectives: A clinicopathologic study with an emphasis on tumor immunohistochemical profile is presented.
Methods: Sixty-one cases of male invasive breast cancers were studied. Median age of the cohort was 65 years.
Results: Ninety-seven percent were estrogen receptor positive+ and 10% human epidermal growth factor receptor 2 positive. The individual diagnostic marker positivity was 98% for GATA-binding protein 3, 95% for androgen receptor, 90% for progesterone receptor, 88% for deleted in pancreatic cancer 4, 75% for gross cystic disease fluid protein 15, 72% for cytokeratin 7, 55% for mammaglobin, and 15% for vimentin and Wilms tumor protein 1. Caudal type homeobox 2 protein, cytokeratin 20, Napsin A, paired box gene 8, prostate-specific antigen, thyroid transcription factor 1, and uroplakin II were negative in all cases. Survival analyses showed tumor stage, receptor status, and Nottingham prognostic index to be prognostic. The overall survival was 70%, but the breast cancer–specific survival was 92% (mean follow-up, 59 months); 33% developed second malignancy. The immunohistochemistry profile was similar to female breast cancers.
Conclusions: The second malignancies in this cohort affected overall survival and suggest the possibility of other germline mutations in addition to BRCA2 in male patients with breast cancer.
Upon completion of this activity you will be able to:
discuss the prognostic/predictive marker profile of male breast cancer.
apply immunohistochemical stains judiciously in identifying male breast cancer.
describe the common morphologic types of male breast cancer.
The ASCP is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians. The ASCP designates this journal-based CME activity for a maximum of 1 AMA PRA Category 1 Credit™ per article. Physicians should claim only the credit commensurate with the extent of their participation in the activity. This activity qualifies as an American Board of Pathology Maintenance of Certification Part II Self-Assessment Module.
The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Exam is located at www.ascp.org/ajcpcme.
Invasive breast carcinoma affected more than 2,000 men in the United States in 2015, with an increasing incidence over the course of the past 4 decades.1‐3 The etiology of male breast cancer is poorly understood, but apart from known genetic risk, incidence is associated with obesity/high body mass index, Klinefelter syndrome, gynecomastia, liver disease, testicular conditions (orchitis, testicular injury or orchiectomy, undescended testicles), alcoholism, and radiation exposure.3‐5 The prevailing thought is that male breast carcinoma has many similarities to postmenopausal female breast cancer but presents at a higher stage and in an older population.6 Additional distinctions between male and female breast cancer have been demonstrated.7 In comparison to female breast cancers, male breast cancers show higher rates of estrogen and progesterone receptor positivity and lower rates of human epidermal growth factor receptor 2 (HER2) positivity, as well as an increased propensity to genetic predisposition.5,8‐10 Male breast cancer has also been shown to be molecularly distinct, with differences in gene expression and methylation patterns, relative risk from cancer predisposition genes such as BRCA2, and unique genetic aberrations.11‐16 Despite the demonstration of these differences, there has been very little focus on establishing an extended immunoprofile of male breast cancers and comparing this immunophenotype with that seen in female breast cancers. The most extensive panel of immunohistochemical stains was published by Kornegoor et al,14 focusing on the expression of oncogenes and tumor suppressors shown to be prognostic in female breast cancer. However, the results of immunohistochemical testing for commonly employed diagnostic markers have not been systematically studied.
Here we present a detailed clinicopathologic study of male breast carcinoma, including an extended immunohistochemical profile.
Materials and Methods
Cases of invasive male breast cancer diagnosed at our institution from January 1, 2003, through December 31, 2013, were retrospectively identified through review of the electronic medical record. The study was approved by the institutional review board at the University of Pittsburgh. Tumor size, patient age at diagnosis, and tumor stage were available from reports. Slides for all cases were reviewed for Nottingham tumor grade. Nottingham prognostic index (NPI), which combines tumor size, lymph node status, and tumor grade into one index,17 was calculated and analyzed with respect to survival.
Areas for tissue microarray (TMA) construction were marked at the time of slide review. A TMA was constructed from formalin-fixed, paraffin-embedded tumor tissues using 0.6-mm cores on a manual microarrayer, MTA1 (Beecher Instruments, Sun Prairie, WI). For immunohistochemical characterization, a panel of diagnostic immunohistochemical stains was performed on TMAs of invasive carcinomas constructed with 3-fold redundancy (ie, three tissue cores per case represented on the array). The diagnostic immunohistochemical panel was chosen to include breast cancer–specific markers and markers that are expressed in tumors resembling breast cancer. The diagnostic panel included androgen receptor (AR), caudal type homeobox 2 protein (CDX2), cytokeratin 7 (CK7), cytokeratin 20 (CK20), deleted in pancreatic cancer 4 (DPC4), GATA-binding protein 3 (GATA3), gross cystic disease fluid protein 15 (GCDFP-15), mammaglobin (MGB), Napsin A, paired box gene 8 (PAX8), prostate-specific antigen (PSA), thyroid transcription factor 1 (TTF1), uroplakin II (UPII), vimentin (VIM), and Wilms tumor protein 1 (WT1). Antigen/antibody details are outlined inTable 1.
Details of Antibodies Used in the Study
| Antibody . | Clone . | Vendor . | Dilution . | Pretreatment . | Detection . | Staining Platform . |
|---|---|---|---|---|---|---|
| AR | SP107 | Ventana | Predilute | CC1, pH 8.5 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| CDX2 | CDX2-88 | BioGenex | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| CK7 | OV-TL 12/30 | Cell Marque | Predilute | Protease 1 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| CK20 | SP33 | Ventana Medical | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| DPC4 | B8 | Santa Cruz Biotechnology | 1:500 | ER2, pH 9.0 | BOND Polymer Refine; Leica | Leica Bond III |
| ER | 6F11 or SP1 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark XT; Ventana |
| GATA3 | L50-823 | Biocare Medical | 1:800 | CC1 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| GCDFP-15 | 23A3 | Leica | Predilute | ER1, pH 6.0; Leica | BOND Polymer Refine; Leica | BOND MAX; Leica |
| HER2 | CB11 or 4B5 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark XT; Ventana |
| MGB | 304-1A5 and 31A5 | Zeta Corporation | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| Napsin A | mRQ-60 | Cell Marque | Predilute | CC1, pH 8.5 | Ultraview/DAB; Ventana | BenchMark Ultra; Ventana |
| PAX8 | Polyclonal | Cell Marque | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| PR | 1A6 or 1E2 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| PSA | Polyclonal | Ventana | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| TTF1 | 8G7G3/1 | Ventana | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| UPII | BC21 | Biocare Medical | Predilute | CC1, pH 8.5 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| VIM | V9 | Leica | Predilute | ER1, pH 6.0 | BOND Polymer Refine; Leica | Leica BOND III |
| WT1 | WT49 | Leica | Predilute | ER2, pH 9.0 | BOND Polymer Refine; Leica | Leica BOND III |
| Antibody . | Clone . | Vendor . | Dilution . | Pretreatment . | Detection . | Staining Platform . |
|---|---|---|---|---|---|---|
| AR | SP107 | Ventana | Predilute | CC1, pH 8.5 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| CDX2 | CDX2-88 | BioGenex | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| CK7 | OV-TL 12/30 | Cell Marque | Predilute | Protease 1 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| CK20 | SP33 | Ventana Medical | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| DPC4 | B8 | Santa Cruz Biotechnology | 1:500 | ER2, pH 9.0 | BOND Polymer Refine; Leica | Leica Bond III |
| ER | 6F11 or SP1 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark XT; Ventana |
| GATA3 | L50-823 | Biocare Medical | 1:800 | CC1 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| GCDFP-15 | 23A3 | Leica | Predilute | ER1, pH 6.0; Leica | BOND Polymer Refine; Leica | BOND MAX; Leica |
| HER2 | CB11 or 4B5 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark XT; Ventana |
| MGB | 304-1A5 and 31A5 | Zeta Corporation | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| Napsin A | mRQ-60 | Cell Marque | Predilute | CC1, pH 8.5 | Ultraview/DAB; Ventana | BenchMark Ultra; Ventana |
| PAX8 | Polyclonal | Cell Marque | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| PR | 1A6 or 1E2 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| PSA | Polyclonal | Ventana | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| TTF1 | 8G7G3/1 | Ventana | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| UPII | BC21 | Biocare Medical | Predilute | CC1, pH 8.5 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| VIM | V9 | Leica | Predilute | ER1, pH 6.0 | BOND Polymer Refine; Leica | Leica BOND III |
| WT1 | WT49 | Leica | Predilute | ER2, pH 9.0 | BOND Polymer Refine; Leica | Leica BOND III |
AR, androgen receptor; CDX2, caudal type homeobox 2 protein; CK7, cytokeratin 7; CK20, cytokeratin 20; DPC4, deleted in pancreatic cancer 4 (also known as SMAD4); ER, estrogen receptor; GATA3, GATA-binding protein 3; GCDFP-15, gross cystic disease fluid protein 15; HER2, human epidermal growth factor receptor 2; MGB, mammaglobin; PAX8, paired box gene 8; PR, progesterone receptor; PSA, prostate-specific antigen; TTF1, thyroid transcription factor 1; UPII, uroplakin II; VIM, vimentin; WT1, Wilms tumor protein 1.
aVendors: Biocare Medical (Concord, CA), BioGenex (Fremont, CA), Cell Marque (Rocklin, CA), Dako Corporation (Carpinteria, CA), Leica Microsystems. (Buffalo Grove, IL), Santa Cruz Biotechnology (Dallas, TX), Ventana Medical Systems (Tucson, AZ), and Zeta Corporation (Sierra Madre, CA).
Details of Antibodies Used in the Study
| Antibody . | Clone . | Vendor . | Dilution . | Pretreatment . | Detection . | Staining Platform . |
|---|---|---|---|---|---|---|
| AR | SP107 | Ventana | Predilute | CC1, pH 8.5 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| CDX2 | CDX2-88 | BioGenex | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| CK7 | OV-TL 12/30 | Cell Marque | Predilute | Protease 1 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| CK20 | SP33 | Ventana Medical | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| DPC4 | B8 | Santa Cruz Biotechnology | 1:500 | ER2, pH 9.0 | BOND Polymer Refine; Leica | Leica Bond III |
| ER | 6F11 or SP1 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark XT; Ventana |
| GATA3 | L50-823 | Biocare Medical | 1:800 | CC1 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| GCDFP-15 | 23A3 | Leica | Predilute | ER1, pH 6.0; Leica | BOND Polymer Refine; Leica | BOND MAX; Leica |
| HER2 | CB11 or 4B5 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark XT; Ventana |
| MGB | 304-1A5 and 31A5 | Zeta Corporation | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| Napsin A | mRQ-60 | Cell Marque | Predilute | CC1, pH 8.5 | Ultraview/DAB; Ventana | BenchMark Ultra; Ventana |
| PAX8 | Polyclonal | Cell Marque | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| PR | 1A6 or 1E2 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| PSA | Polyclonal | Ventana | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| TTF1 | 8G7G3/1 | Ventana | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| UPII | BC21 | Biocare Medical | Predilute | CC1, pH 8.5 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| VIM | V9 | Leica | Predilute | ER1, pH 6.0 | BOND Polymer Refine; Leica | Leica BOND III |
| WT1 | WT49 | Leica | Predilute | ER2, pH 9.0 | BOND Polymer Refine; Leica | Leica BOND III |
| Antibody . | Clone . | Vendor . | Dilution . | Pretreatment . | Detection . | Staining Platform . |
|---|---|---|---|---|---|---|
| AR | SP107 | Ventana | Predilute | CC1, pH 8.5 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| CDX2 | CDX2-88 | BioGenex | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| CK7 | OV-TL 12/30 | Cell Marque | Predilute | Protease 1 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| CK20 | SP33 | Ventana Medical | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| DPC4 | B8 | Santa Cruz Biotechnology | 1:500 | ER2, pH 9.0 | BOND Polymer Refine; Leica | Leica Bond III |
| ER | 6F11 or SP1 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark XT; Ventana |
| GATA3 | L50-823 | Biocare Medical | 1:800 | CC1 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| GCDFP-15 | 23A3 | Leica | Predilute | ER1, pH 6.0; Leica | BOND Polymer Refine; Leica | BOND MAX; Leica |
| HER2 | CB11 or 4B5 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark XT; Ventana |
| MGB | 304-1A5 and 31A5 | Zeta Corporation | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| Napsin A | mRQ-60 | Cell Marque | Predilute | CC1, pH 8.5 | Ultraview/DAB; Ventana | BenchMark Ultra; Ventana |
| PAX8 | Polyclonal | Cell Marque | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| PR | 1A6 or 1E2 | Ventana | Predilute | CC1, pH 8.5 | iView/DAB; Ventana | BenchMark Ultra; Ventana |
| PSA | Polyclonal | Ventana | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| TTF1 | 8G7G3/1 | Ventana | Predilute | CC1, pH 8.5 | UltraView/DAB; Ventana | BenchMark Ultra; Ventana |
| UPII | BC21 | Biocare Medical | Predilute | CC1, pH 8.5 | OptiView/DAB; Ventana | BenchMark Ultra; Ventana |
| VIM | V9 | Leica | Predilute | ER1, pH 6.0 | BOND Polymer Refine; Leica | Leica BOND III |
| WT1 | WT49 | Leica | Predilute | ER2, pH 9.0 | BOND Polymer Refine; Leica | Leica BOND III |
AR, androgen receptor; CDX2, caudal type homeobox 2 protein; CK7, cytokeratin 7; CK20, cytokeratin 20; DPC4, deleted in pancreatic cancer 4 (also known as SMAD4); ER, estrogen receptor; GATA3, GATA-binding protein 3; GCDFP-15, gross cystic disease fluid protein 15; HER2, human epidermal growth factor receptor 2; MGB, mammaglobin; PAX8, paired box gene 8; PR, progesterone receptor; PSA, prostate-specific antigen; TTF1, thyroid transcription factor 1; UPII, uroplakin II; VIM, vimentin; WT1, Wilms tumor protein 1.
aVendors: Biocare Medical (Concord, CA), BioGenex (Fremont, CA), Cell Marque (Rocklin, CA), Dako Corporation (Carpinteria, CA), Leica Microsystems. (Buffalo Grove, IL), Santa Cruz Biotechnology (Dallas, TX), Ventana Medical Systems (Tucson, AZ), and Zeta Corporation (Sierra Madre, CA).
All markers were scored using the semiquantitative H-score method, which is calculated as the sum of the percent staining multiplied by an ordinal value corresponding to the intensity level (0 = absent, 1 = weak, 2 = moderate, 3 = strong). With four intensity levels, the resulting score ranges from 0 (no staining in the tumor) to 300 (diffuse intense staining of the tumor). An H-score of more than 10 was considered a positive result since focal weak staining is not diagnostically useful in the workup of tumors of unknown origin. The only exception was GCDFP-15, where the positive cutoff was an H-score of 1 since even focal weak staining (H-score of 1-10) with GCDFP-15 cannot be ignored in the workup of a carcinoma of unknown origin.18
Results of estrogen receptor (ER), progesterone receptor (PR), HER2, and Ki-67 were obtained from pathology reports. For those few cases with missing immunohistochemical results for ER, PR, HER2, and Ki-67, stains were performed on whole tissue sections. ER/PR results were classified as positive or negative using the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines19 and were also semiquantified using the H-score method. HER2 results were categorized using 2013 ASCO/CAP guidelines,20 that is, HER2 immunohistochemical scores of 0 and 1+ considered as negative, score of 3+ as positive, and score of 2+ tested by fluorescence in situ hybridization with further classification into negative, equivocal, and positive performed using 2013 ASCO/CAP in situ hybridization criteria. These results allowed for classification of tumors into four categories (ER+/HER2–, ER+/HER2+, ER–/HER2+, ER–/HER2–), reflecting surrogates for different molecular categories. HER2-equivocal tumors were included with HER2– for categorization. This categorization allowed correlation between the diagnostic markers and the most well-known prognostic/predictive markers of breast cancer.
Treatment and clinical follow-up were obtained from tumor registry data. Disease-free survival (DFS) was defined as time from diagnosis to development of any recurrence (distant or locoregional) or last follow-up. Distant disease-free survival (DDFS) was defined as time from diagnosis to development of distant recurrence or last follow-up. Overall survival (OS) duration was defined as the time from diagnosis to death from any cause or last follow-up. Breast cancer–specific survival (BCSS) duration was defined as the time from diagnosis to death due to breast cancer or last follow-up.
The morphologic and immunohistochemical results were also compared with a preexisting data set of 198 consecutive female breast cancers.21 Statistical analysis regarding association of diagnostic markers with clinicopathologic factors, as well as diagnostic marker comparison between male and female breast cancers, was performed using GraphPad QuickCalcs online software (GraphPad Software, La Jolla, CA). For comparison of means, independent sample t tests were performed. Univariate analysis was performed using χ2 and Fisher exact tests to compare the differences in percentages between groups. A P value less than .05 was considered significant.
Multiple morphologic, clinical, and immunohistochemical markers were individually analyzed by means of the log-rank test to assess their effect on survival. Statistical analyses for survival were performed using STATA 12.0 (StataCorp, College Station, TX) and SPSS 20.0 (SPSS, Chicago, IL). Only univariate analysis was performed. Multivariate analysis could not be performed due to a lower number of total cases.
Results
Patient and Tumor Characteristics
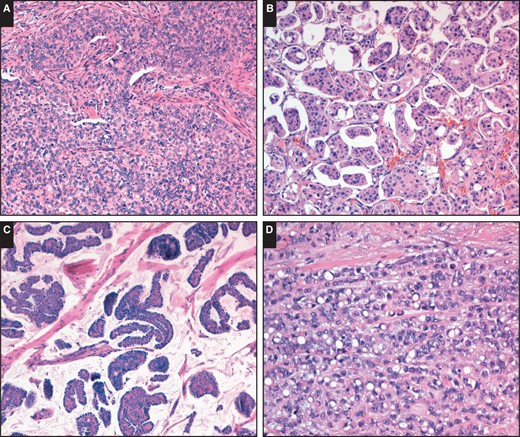
Although most carcinomas were ductal, no special type, there was a trend toward a solid-papillary growth pattern (A; H&E, ×100). Other tumor types were also noted. B, Micropapillary growth pattern (H&E, ×100). C, Mucinous carcinoma (H&E, ×100). D, Pleomorphic lobular carcinoma with signet ring cells (H&E, ×200).
Diagnostic Marker Results
Due to tissue loss in some cases, diagnostic marker results were not available on all cases. The individual diagnostic marker positivity in decreasing order was 98% (54/55) for GATA3, 95% (52/55) for AR, 90% (54/60) for PR, 88% (46/52) for DPC4, 75% (41/55) for GCDFP-15, 72% (39/54) for CK7, 55% (30/55) for MGB, 15% (8/54) for VIM, and 15% (8/55) for WT1. Seven markers were negative for all cases, including CDX 2 (0/53), CK20 (0/54), Napsin A (0/55), PAX8 (0/55), PSA (0/54), TTF1 (0/55), and UPII (0/54), as shown inTable 2. Data for all markers were available for 50 carcinomas (44 ER+/HER2–, four ER+/HER2+, one ER–/HER2+, and one ER–/HER2–), resulting in a total of 28 different profiles. The most common profile was ER+/HER2–/AR+/CDX2–/CK7+/CK20–/DPC4+/GATA3+/GCDFP-15+/MGB+/Napsin A–/PAX8–/PR+/PSA–/TTF1–/UPII–/VIM–/WT1–.
Individual Marker Reactivity in Different "Molecular" Types of Breast Cancers and in All Tumors Combineda
| Antigen . | ER+/HER2– . | ER+/HER2+ . | ER–/HER2+ . | ER–/HER2– . | All Tumors . |
|---|---|---|---|---|---|
| AR | |||||
| No. (%) | 47/49 (96) | 4/4 (100) | 1/1 (100) | 0/1 (0) | 52/55 (95) |
| Mean | 227 | 95 | 240 | NA | 228 |
| Median | 260 | 95 | 240 | NA | 255 |
| Range | 30-300 | 90-100 | NA | NA | 30-300 |
| CDX2, No. (%) | 0/47 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/53 (0) |
| CK7 | |||||
| No. (%) | 34/48 (71) | 3/4 (75) | 1/1 (100) | 1/1 (100) | 39/54 (72) |
| Mean | 272 | 267 | 300 | 300 | 273 |
| Median | 300 | 300 | 300 | 300 | 300 |
| Range | 50-300 | 200-300 | NA | NA | 50-300 |
| CK20, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| DPC4 | |||||
| No. (%) | 41/46 (89) | 4/4 (100) | 0/1 (0) | 1/1 (100) | 46/52 (88) |
| Mean | 133 | 153 | NA | 100 | 134 |
| Median | 120 | 155 | NA | 100 | 135 |
| Range | 20-270 | 100-160 | NA | NA | 20-270 |
| GATA3 | |||||
| No. (%) | 49/49 (100) | 4/4 (100) | 1/1 (100) | 0/1 (0) | 54/55 (98) |
| Mean | 248 | 253 | 80 | NA | 245 |
| Median | 270 | 255 | 80 | NA | 270 |
| Range | 70-300 | 220-280 | NA | NA | 70-300 |
| GCDFP-15 | |||||
| No. (%) | 35/49 (71) | 4/4 (100) | 1/1 (100) | 1/1 (100) | 41/55 (75) |
| Mean | 97 | 57 | 140 | 3 | 92 |
| Median | 70 | 55 | 140 | 3 | 70 |
| Range | 1-300 | 9-110 | NA | NA | 1-300 |
| MGB | |||||
| No. (%) | 27/49 (55) | 3/4 (75) | 0/1 (0) | 0/1 (0) | 30/55 (55) |
| Mean | 128 | 130 | NA | NA | 128 |
| Median | 100 | 55 | NA | NA | 95 |
| Range | 11-300 | 35-300 | NA | NA | 11-300 |
| Napsin A, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| PAX8, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| PR | |||||
| No. (%) | 50/53 (94) | 4/5 (80) | 0/1 (0) | 0/1 (0) | 54/60 (90) |
| Mean | 153 | 106 | NA | NA | 150 |
| Median | 170 | 68 | NA | NA | 165 |
| Range | 3-300 | 3-285 | NA | NA | 3-300 |
| PSA, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| TTF1, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| UPII, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| VIM | |||||
| No. (%) | 6/48 (13) | 2/4 (50) | 0/1 (0) | 0/1 (0) | 8/54 (15) |
| Mean | 97 | 68 | NA | NA | 89 |
| Median | 75 | 68 | NA | NA | 75 |
| Range | 15-220 | 15-120 | NA | NA | 15-220 |
| WT1 | |||||
| No. (%) | 7/49 (14) | 1/4 (25) | 0/1 (0) | 0/1 (0) | 8/55 (15) |
| Mean | 81 | 80b | NA | NA | 81 |
| Median | 20 | NA | NA | 33 | |
| Range | 15-270 | NA | NA | 15-270 |
| Antigen . | ER+/HER2– . | ER+/HER2+ . | ER–/HER2+ . | ER–/HER2– . | All Tumors . |
|---|---|---|---|---|---|
| AR | |||||
| No. (%) | 47/49 (96) | 4/4 (100) | 1/1 (100) | 0/1 (0) | 52/55 (95) |
| Mean | 227 | 95 | 240 | NA | 228 |
| Median | 260 | 95 | 240 | NA | 255 |
| Range | 30-300 | 90-100 | NA | NA | 30-300 |
| CDX2, No. (%) | 0/47 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/53 (0) |
| CK7 | |||||
| No. (%) | 34/48 (71) | 3/4 (75) | 1/1 (100) | 1/1 (100) | 39/54 (72) |
| Mean | 272 | 267 | 300 | 300 | 273 |
| Median | 300 | 300 | 300 | 300 | 300 |
| Range | 50-300 | 200-300 | NA | NA | 50-300 |
| CK20, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| DPC4 | |||||
| No. (%) | 41/46 (89) | 4/4 (100) | 0/1 (0) | 1/1 (100) | 46/52 (88) |
| Mean | 133 | 153 | NA | 100 | 134 |
| Median | 120 | 155 | NA | 100 | 135 |
| Range | 20-270 | 100-160 | NA | NA | 20-270 |
| GATA3 | |||||
| No. (%) | 49/49 (100) | 4/4 (100) | 1/1 (100) | 0/1 (0) | 54/55 (98) |
| Mean | 248 | 253 | 80 | NA | 245 |
| Median | 270 | 255 | 80 | NA | 270 |
| Range | 70-300 | 220-280 | NA | NA | 70-300 |
| GCDFP-15 | |||||
| No. (%) | 35/49 (71) | 4/4 (100) | 1/1 (100) | 1/1 (100) | 41/55 (75) |
| Mean | 97 | 57 | 140 | 3 | 92 |
| Median | 70 | 55 | 140 | 3 | 70 |
| Range | 1-300 | 9-110 | NA | NA | 1-300 |
| MGB | |||||
| No. (%) | 27/49 (55) | 3/4 (75) | 0/1 (0) | 0/1 (0) | 30/55 (55) |
| Mean | 128 | 130 | NA | NA | 128 |
| Median | 100 | 55 | NA | NA | 95 |
| Range | 11-300 | 35-300 | NA | NA | 11-300 |
| Napsin A, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| PAX8, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| PR | |||||
| No. (%) | 50/53 (94) | 4/5 (80) | 0/1 (0) | 0/1 (0) | 54/60 (90) |
| Mean | 153 | 106 | NA | NA | 150 |
| Median | 170 | 68 | NA | NA | 165 |
| Range | 3-300 | 3-285 | NA | NA | 3-300 |
| PSA, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| TTF1, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| UPII, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| VIM | |||||
| No. (%) | 6/48 (13) | 2/4 (50) | 0/1 (0) | 0/1 (0) | 8/54 (15) |
| Mean | 97 | 68 | NA | NA | 89 |
| Median | 75 | 68 | NA | NA | 75 |
| Range | 15-220 | 15-120 | NA | NA | 15-220 |
| WT1 | |||||
| No. (%) | 7/49 (14) | 1/4 (25) | 0/1 (0) | 0/1 (0) | 8/55 (15) |
| Mean | 81 | 80b | NA | NA | 81 |
| Median | 20 | NA | NA | 33 | |
| Range | 15-270 | NA | NA | 15-270 |
AR, androgen receptor; CDX2, caudal type homeobox 2 protein; CK7, cytokeratin 7; CK20, cytokeratin 20; DPC4, deleted in pancreatic cancer 4 (also known as SMAD4); ER, estrogen receptor; GATA3, GATA-binding protein 3; GCDFP-15, gross cystic disease fluid protein 15; HER2, human epidermal growth factor receptor 2; MGB, mammaglobin; NA, not applicable; PAX8, paired box gene 8; PR, progesterone receptor; PSA, prostate-specific antigen; TTF1, thyroid transcription factor 1; UPII, uroplakin II; VIM, vimentin; WT1, Wilms tumor protein 1.
aPositive for all markers (except ER, PR, and GCDFP-15) defined as H-score more than 10. For ER, PR, and GCDFP-15, positive is H-score of 1 or more. Mean, median, and range calculation is for H-scores and excludes negative cases.
bH-score.
Individual Marker Reactivity in Different "Molecular" Types of Breast Cancers and in All Tumors Combineda
| Antigen . | ER+/HER2– . | ER+/HER2+ . | ER–/HER2+ . | ER–/HER2– . | All Tumors . |
|---|---|---|---|---|---|
| AR | |||||
| No. (%) | 47/49 (96) | 4/4 (100) | 1/1 (100) | 0/1 (0) | 52/55 (95) |
| Mean | 227 | 95 | 240 | NA | 228 |
| Median | 260 | 95 | 240 | NA | 255 |
| Range | 30-300 | 90-100 | NA | NA | 30-300 |
| CDX2, No. (%) | 0/47 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/53 (0) |
| CK7 | |||||
| No. (%) | 34/48 (71) | 3/4 (75) | 1/1 (100) | 1/1 (100) | 39/54 (72) |
| Mean | 272 | 267 | 300 | 300 | 273 |
| Median | 300 | 300 | 300 | 300 | 300 |
| Range | 50-300 | 200-300 | NA | NA | 50-300 |
| CK20, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| DPC4 | |||||
| No. (%) | 41/46 (89) | 4/4 (100) | 0/1 (0) | 1/1 (100) | 46/52 (88) |
| Mean | 133 | 153 | NA | 100 | 134 |
| Median | 120 | 155 | NA | 100 | 135 |
| Range | 20-270 | 100-160 | NA | NA | 20-270 |
| GATA3 | |||||
| No. (%) | 49/49 (100) | 4/4 (100) | 1/1 (100) | 0/1 (0) | 54/55 (98) |
| Mean | 248 | 253 | 80 | NA | 245 |
| Median | 270 | 255 | 80 | NA | 270 |
| Range | 70-300 | 220-280 | NA | NA | 70-300 |
| GCDFP-15 | |||||
| No. (%) | 35/49 (71) | 4/4 (100) | 1/1 (100) | 1/1 (100) | 41/55 (75) |
| Mean | 97 | 57 | 140 | 3 | 92 |
| Median | 70 | 55 | 140 | 3 | 70 |
| Range | 1-300 | 9-110 | NA | NA | 1-300 |
| MGB | |||||
| No. (%) | 27/49 (55) | 3/4 (75) | 0/1 (0) | 0/1 (0) | 30/55 (55) |
| Mean | 128 | 130 | NA | NA | 128 |
| Median | 100 | 55 | NA | NA | 95 |
| Range | 11-300 | 35-300 | NA | NA | 11-300 |
| Napsin A, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| PAX8, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| PR | |||||
| No. (%) | 50/53 (94) | 4/5 (80) | 0/1 (0) | 0/1 (0) | 54/60 (90) |
| Mean | 153 | 106 | NA | NA | 150 |
| Median | 170 | 68 | NA | NA | 165 |
| Range | 3-300 | 3-285 | NA | NA | 3-300 |
| PSA, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| TTF1, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| UPII, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| VIM | |||||
| No. (%) | 6/48 (13) | 2/4 (50) | 0/1 (0) | 0/1 (0) | 8/54 (15) |
| Mean | 97 | 68 | NA | NA | 89 |
| Median | 75 | 68 | NA | NA | 75 |
| Range | 15-220 | 15-120 | NA | NA | 15-220 |
| WT1 | |||||
| No. (%) | 7/49 (14) | 1/4 (25) | 0/1 (0) | 0/1 (0) | 8/55 (15) |
| Mean | 81 | 80b | NA | NA | 81 |
| Median | 20 | NA | NA | 33 | |
| Range | 15-270 | NA | NA | 15-270 |
| Antigen . | ER+/HER2– . | ER+/HER2+ . | ER–/HER2+ . | ER–/HER2– . | All Tumors . |
|---|---|---|---|---|---|
| AR | |||||
| No. (%) | 47/49 (96) | 4/4 (100) | 1/1 (100) | 0/1 (0) | 52/55 (95) |
| Mean | 227 | 95 | 240 | NA | 228 |
| Median | 260 | 95 | 240 | NA | 255 |
| Range | 30-300 | 90-100 | NA | NA | 30-300 |
| CDX2, No. (%) | 0/47 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/53 (0) |
| CK7 | |||||
| No. (%) | 34/48 (71) | 3/4 (75) | 1/1 (100) | 1/1 (100) | 39/54 (72) |
| Mean | 272 | 267 | 300 | 300 | 273 |
| Median | 300 | 300 | 300 | 300 | 300 |
| Range | 50-300 | 200-300 | NA | NA | 50-300 |
| CK20, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| DPC4 | |||||
| No. (%) | 41/46 (89) | 4/4 (100) | 0/1 (0) | 1/1 (100) | 46/52 (88) |
| Mean | 133 | 153 | NA | 100 | 134 |
| Median | 120 | 155 | NA | 100 | 135 |
| Range | 20-270 | 100-160 | NA | NA | 20-270 |
| GATA3 | |||||
| No. (%) | 49/49 (100) | 4/4 (100) | 1/1 (100) | 0/1 (0) | 54/55 (98) |
| Mean | 248 | 253 | 80 | NA | 245 |
| Median | 270 | 255 | 80 | NA | 270 |
| Range | 70-300 | 220-280 | NA | NA | 70-300 |
| GCDFP-15 | |||||
| No. (%) | 35/49 (71) | 4/4 (100) | 1/1 (100) | 1/1 (100) | 41/55 (75) |
| Mean | 97 | 57 | 140 | 3 | 92 |
| Median | 70 | 55 | 140 | 3 | 70 |
| Range | 1-300 | 9-110 | NA | NA | 1-300 |
| MGB | |||||
| No. (%) | 27/49 (55) | 3/4 (75) | 0/1 (0) | 0/1 (0) | 30/55 (55) |
| Mean | 128 | 130 | NA | NA | 128 |
| Median | 100 | 55 | NA | NA | 95 |
| Range | 11-300 | 35-300 | NA | NA | 11-300 |
| Napsin A, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| PAX8, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| PR | |||||
| No. (%) | 50/53 (94) | 4/5 (80) | 0/1 (0) | 0/1 (0) | 54/60 (90) |
| Mean | 153 | 106 | NA | NA | 150 |
| Median | 170 | 68 | NA | NA | 165 |
| Range | 3-300 | 3-285 | NA | NA | 3-300 |
| PSA, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| TTF1, No. (%) | 0/49 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/55 (0) |
| UPII, No. (%) | 0/48 (0) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/54 (0) |
| VIM | |||||
| No. (%) | 6/48 (13) | 2/4 (50) | 0/1 (0) | 0/1 (0) | 8/54 (15) |
| Mean | 97 | 68 | NA | NA | 89 |
| Median | 75 | 68 | NA | NA | 75 |
| Range | 15-220 | 15-120 | NA | NA | 15-220 |
| WT1 | |||||
| No. (%) | 7/49 (14) | 1/4 (25) | 0/1 (0) | 0/1 (0) | 8/55 (15) |
| Mean | 81 | 80b | NA | NA | 81 |
| Median | 20 | NA | NA | 33 | |
| Range | 15-270 | NA | NA | 15-270 |
AR, androgen receptor; CDX2, caudal type homeobox 2 protein; CK7, cytokeratin 7; CK20, cytokeratin 20; DPC4, deleted in pancreatic cancer 4 (also known as SMAD4); ER, estrogen receptor; GATA3, GATA-binding protein 3; GCDFP-15, gross cystic disease fluid protein 15; HER2, human epidermal growth factor receptor 2; MGB, mammaglobin; NA, not applicable; PAX8, paired box gene 8; PR, progesterone receptor; PSA, prostate-specific antigen; TTF1, thyroid transcription factor 1; UPII, uroplakin II; VIM, vimentin; WT1, Wilms tumor protein 1.
aPositive for all markers (except ER, PR, and GCDFP-15) defined as H-score more than 10. For ER, PR, and GCDFP-15, positive is H-score of 1 or more. Mean, median, and range calculation is for H-scores and excludes negative cases.
bH-score.
Univariate analysis was performed for each diagnostic marker vs clinicopathologic factors. Statistically significant associations are shown in Table 3.
Statistical Association and Between Diagnostic Markers and Clinicopathologic Featuresa
| Diagnostic Marker . | Clinicopathologic Associations of Interest . |
|---|---|
| Androgen receptor | No statistically significant associations |
| Caudal type homeobox 2 protein | Analysis not performed (reason: all cases negative) |
| Cytokeratin 7 (CK7) |
|
| Cytokeratin 20 | Analysis not performed (reason: all cases negative) |
| Deleted in pancreatic cancer 4 (DPC4, also known as SMAD4) |
|
| GATA binding protein 3 | Analysis not performed (reason: only one case negative) |
| Gross cystic disease fluid protein 15 (GCDFP-15) |
|
| Mammaglobin | No statistically significant associations |
| Napsin A | Analysis not performed (reason: all cases negative) |
| Paired box gene 8 | Analysis not performed (reason: all cases negative) |
| Progesterone receptor (PR) |
|
| Prostate-specific antigen | Analysis not performed (reason: all cases negative) |
| Thyroid transcription factor 1 | Analysis not performed (reason: all cases negative) |
| Uroplakin II | Analysis not performed (reason: all cases negative) |
| Vimentin | No statistically significant associations |
| Wilms tumor protein 1 | No statistically significant associations |
| Diagnostic Marker . | Clinicopathologic Associations of Interest . |
|---|---|
| Androgen receptor | No statistically significant associations |
| Caudal type homeobox 2 protein | Analysis not performed (reason: all cases negative) |
| Cytokeratin 7 (CK7) |
|
| Cytokeratin 20 | Analysis not performed (reason: all cases negative) |
| Deleted in pancreatic cancer 4 (DPC4, also known as SMAD4) |
|
| GATA binding protein 3 | Analysis not performed (reason: only one case negative) |
| Gross cystic disease fluid protein 15 (GCDFP-15) |
|
| Mammaglobin | No statistically significant associations |
| Napsin A | Analysis not performed (reason: all cases negative) |
| Paired box gene 8 | Analysis not performed (reason: all cases negative) |
| Progesterone receptor (PR) |
|
| Prostate-specific antigen | Analysis not performed (reason: all cases negative) |
| Thyroid transcription factor 1 | Analysis not performed (reason: all cases negative) |
| Uroplakin II | Analysis not performed (reason: all cases negative) |
| Vimentin | No statistically significant associations |
| Wilms tumor protein 1 | No statistically significant associations |
aP values of less than .05 were considered significant. Larger P values (.05 to < 0.1) were not considered significant but could give a clue to a “trend.”
Statistical Association and Between Diagnostic Markers and Clinicopathologic Featuresa
| Diagnostic Marker . | Clinicopathologic Associations of Interest . |
|---|---|
| Androgen receptor | No statistically significant associations |
| Caudal type homeobox 2 protein | Analysis not performed (reason: all cases negative) |
| Cytokeratin 7 (CK7) |
|
| Cytokeratin 20 | Analysis not performed (reason: all cases negative) |
| Deleted in pancreatic cancer 4 (DPC4, also known as SMAD4) |
|
| GATA binding protein 3 | Analysis not performed (reason: only one case negative) |
| Gross cystic disease fluid protein 15 (GCDFP-15) |
|
| Mammaglobin | No statistically significant associations |
| Napsin A | Analysis not performed (reason: all cases negative) |
| Paired box gene 8 | Analysis not performed (reason: all cases negative) |
| Progesterone receptor (PR) |
|
| Prostate-specific antigen | Analysis not performed (reason: all cases negative) |
| Thyroid transcription factor 1 | Analysis not performed (reason: all cases negative) |
| Uroplakin II | Analysis not performed (reason: all cases negative) |
| Vimentin | No statistically significant associations |
| Wilms tumor protein 1 | No statistically significant associations |
| Diagnostic Marker . | Clinicopathologic Associations of Interest . |
|---|---|
| Androgen receptor | No statistically significant associations |
| Caudal type homeobox 2 protein | Analysis not performed (reason: all cases negative) |
| Cytokeratin 7 (CK7) |
|
| Cytokeratin 20 | Analysis not performed (reason: all cases negative) |
| Deleted in pancreatic cancer 4 (DPC4, also known as SMAD4) |
|
| GATA binding protein 3 | Analysis not performed (reason: only one case negative) |
| Gross cystic disease fluid protein 15 (GCDFP-15) |
|
| Mammaglobin | No statistically significant associations |
| Napsin A | Analysis not performed (reason: all cases negative) |
| Paired box gene 8 | Analysis not performed (reason: all cases negative) |
| Progesterone receptor (PR) |
|
| Prostate-specific antigen | Analysis not performed (reason: all cases negative) |
| Thyroid transcription factor 1 | Analysis not performed (reason: all cases negative) |
| Uroplakin II | Analysis not performed (reason: all cases negative) |
| Vimentin | No statistically significant associations |
| Wilms tumor protein 1 | No statistically significant associations |
aP values of less than .05 were considered significant. Larger P values (.05 to < 0.1) were not considered significant but could give a clue to a “trend.”
Clinicopathologic prognostic variables were compared with a previously published and preexisting data set of 198 consecutive female breast cancers.21 The results are detailed in Table 4. With respect to diagnostic immunohistochemistry (IHC) markers, significantly higher numbers of male breast cancer cases were positive for AR, PR, and GCDFP-15 in comparison to female breast cancers (95% vs 80% for AR with P value of .0080, 90% vs 81% for PR with P value of .01178, and 75% vs 26% for GCDFP-15 with a P value of .0001). Conversely, significantly lower numbers of male breast cancers were positive for CK7 and VIM (72% vs 92% for CK7 with P value of .0002 and 15% vs 31% for VIM with a P value of .0239). No significant differences were identified for staining with CK20, GATA3, MGB, and PAX8.
Comparison of Male Breast Cancers With Preexisting Data Set of Consecutive Female Breast Cancersa
| Patient/Tumor Characteristic . | Male (n = 61) . | Female (n = 198) . | P Value . |
|---|---|---|---|
| Age, mean, y | 64.5 | 60 | .0119b |
| Tumor size, mean, cm | 2.3 | 1.7 | .0002b |
| Ductal cancers | 60/61 (98) | 181/198 (91) | .0821 |
| Nottingham score ≥7 | 39/61 (64) | 93/198 (47) | .0276b |
| Lymph node positive | 29/55 (53) | 62/182 (34) | .0173b |
| ER + | 58/60 (97) | 164/198 (83) | .0051b |
| PR + | 54/60 (90) | 160/198 (81) | .1178 |
| ER H-score (positive cases), mean | 240 | 228 | .2212 |
| PR H-score (positive cases), mean | 150 | 130 | .1803 |
| HER2+ | 6/60 (10) | 26/198 (13) | .6566 |
| Ki-67 index, mean, % | 33c | 25d | .0145b |
| Stage I cases | 12/55 (22) | 119/198 (60) | <.0001b |
| Patient/Tumor Characteristic . | Male (n = 61) . | Female (n = 198) . | P Value . |
|---|---|---|---|
| Age, mean, y | 64.5 | 60 | .0119b |
| Tumor size, mean, cm | 2.3 | 1.7 | .0002b |
| Ductal cancers | 60/61 (98) | 181/198 (91) | .0821 |
| Nottingham score ≥7 | 39/61 (64) | 93/198 (47) | .0276b |
| Lymph node positive | 29/55 (53) | 62/182 (34) | .0173b |
| ER + | 58/60 (97) | 164/198 (83) | .0051b |
| PR + | 54/60 (90) | 160/198 (81) | .1178 |
| ER H-score (positive cases), mean | 240 | 228 | .2212 |
| PR H-score (positive cases), mean | 150 | 130 | .1803 |
| HER2+ | 6/60 (10) | 26/198 (13) | .6566 |
| Ki-67 index, mean, % | 33c | 25d | .0145b |
| Stage I cases | 12/55 (22) | 119/198 (60) | <.0001b |
ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
aValues are presented as number (%) unless otherwise indicated.
bStatistically significant.
cBased on 56 cases.
dBased on 192 cases.
Comparison of Male Breast Cancers With Preexisting Data Set of Consecutive Female Breast Cancersa
| Patient/Tumor Characteristic . | Male (n = 61) . | Female (n = 198) . | P Value . |
|---|---|---|---|
| Age, mean, y | 64.5 | 60 | .0119b |
| Tumor size, mean, cm | 2.3 | 1.7 | .0002b |
| Ductal cancers | 60/61 (98) | 181/198 (91) | .0821 |
| Nottingham score ≥7 | 39/61 (64) | 93/198 (47) | .0276b |
| Lymph node positive | 29/55 (53) | 62/182 (34) | .0173b |
| ER + | 58/60 (97) | 164/198 (83) | .0051b |
| PR + | 54/60 (90) | 160/198 (81) | .1178 |
| ER H-score (positive cases), mean | 240 | 228 | .2212 |
| PR H-score (positive cases), mean | 150 | 130 | .1803 |
| HER2+ | 6/60 (10) | 26/198 (13) | .6566 |
| Ki-67 index, mean, % | 33c | 25d | .0145b |
| Stage I cases | 12/55 (22) | 119/198 (60) | <.0001b |
| Patient/Tumor Characteristic . | Male (n = 61) . | Female (n = 198) . | P Value . |
|---|---|---|---|
| Age, mean, y | 64.5 | 60 | .0119b |
| Tumor size, mean, cm | 2.3 | 1.7 | .0002b |
| Ductal cancers | 60/61 (98) | 181/198 (91) | .0821 |
| Nottingham score ≥7 | 39/61 (64) | 93/198 (47) | .0276b |
| Lymph node positive | 29/55 (53) | 62/182 (34) | .0173b |
| ER + | 58/60 (97) | 164/198 (83) | .0051b |
| PR + | 54/60 (90) | 160/198 (81) | .1178 |
| ER H-score (positive cases), mean | 240 | 228 | .2212 |
| PR H-score (positive cases), mean | 150 | 130 | .1803 |
| HER2+ | 6/60 (10) | 26/198 (13) | .6566 |
| Ki-67 index, mean, % | 33c | 25d | .0145b |
| Stage I cases | 12/55 (22) | 119/198 (60) | <.0001b |
ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
aValues are presented as number (%) unless otherwise indicated.
bStatistically significant.
cBased on 56 cases.
dBased on 192 cases.
Treatment and Survival Data
Clinical follow-up data showed that 77% (40/52, data unknown in nine cases) of patients received adjuvant hormonal therapy, 54% (30/56, data unknown in five cases) received adjuvant chemotherapy, and 32% (17/54, data unknown in seven cases) received adjuvant radiation. The average follow-up time was 59 months (median, 58 months; range, 1-146 months). Nine patients (15%) had a recurrence (eight distant and one locoregional), with an average DFS of 56 months. The OS in the cohort was 70% (43/61). With an average follow-up time of 59 months, this reflects the actual ∼5-year OS rate of 70%. Of the 18 patients who died, cause of death was unknown in two cases. Eleven died of non–breast cancer causes, and only five died of breast cancer, with an actual ∼5-year BCSS rate of 92%. Of the 11 patients who died of non–breast cancer causes, six died of second cancers (esophageal, colon, pancreas, prostate, lung, brain). Second cancers were common in this cohort, seen in 20 (33%) of the 61 patients. Excluding skin cancers, a total of 22 second malignancies developed in 20 patients. These included five prostatic adenocarcinomas, three renal clear cell carcinomas, three lymphomas (two diffuse large B cell and one marginal zone), two lung adenocarcinomas (one usual type and one mucin producing), two thyroid Hürthle cell carcinomas (one Hürthle cell with focal invasion and one papillary carcinoma with Hürthle cell features), two colonic adenocarcinomas, and one each of esophageal adenocarcinoma, bladder urothelial carcinoma, pancreatic adenocarcinoma, parathyroid carcinoma, and glioblastoma. In 13 patients, the second malignancy developed after the diagnosis of male breast carcinoma.
On univariate analysis, improved DFS was associated with administration of radiation (P = .0379) and lower tumor mitotic activity score (P = .0394). Similarly, improved DDFS was associated with lower pathologic nodal status (P = .0189), administration of radiation (P = .0139), lower stage (P = .0345), lower NPI risk category (P = .0358), and lower NPI score (P = .049). For OS, presence of DCIS (P = .0272), GATA3 expression (P < .0001), and ER+/HER2– breast tumor subtype (P < .0001) were significantly associated with survival. Similarly, positive ER status (P = .0006), GATA3 expression (P < .0001), and ER+/HER2– breast tumor subtype (P < .0001) were significantly associated with improved BCSS. With the exception of GATA3, no other diagnostic marker showed any significant association with survival. Genetic testing information for the BRCA2 gene was available for only eight patients, and five (63%) tested positive.
Discussion
Compared with females, breast cancer in males is rare but still affects around 2,000 men per year in the United States. Therefore, it is important to understand clinical and pathologic features of male breast cancer for accurate diagnosis and patient management. Prior studies of male breast cancers have shown that compared with female patients with breast cancer, male patients with breast cancer are slightly older at diagnosis and have a higher tumor stage at presentation.5,8‐10,22 It has also been shown that most male breast cancers are hormone receptor positive. The patient characteristics and tumor features that we report in the current study are very similar to the published data from Surveillance, Epidemiology, and End Results (SEER),23 which analyzed patients diagnosed during the same period of time as our study, indicating that the sample in our study is quite representative of the general population. However, details regarding the relationship between tumor morphology and a broad range of immunohistochemical results are lacking in the literature. The goal of our study was to comprehensively characterize male breast carcinomas morphologically and immunohistochemically.
Similar to female breast cancers, most cases in our cohort of male breast cancers were classified as no special type invasive ductal carcinoma; however, there was a trend toward a papillary growth pattern. There was only one case of invasive lobular carcinoma in this cohort, an expected finding considering the well-documented infrequency of this diagnosis in men. Male breast cancers more often exhibited features associated with a biologically aggressive course, including larger tumor size and higher Nottingham score compared with the preexisting data set of consecutive female breast cancers at our institution. Male breast cancers showed higher proliferative activity, as measured by the Ki-67 proliferation index (mean labeling index of 33%), and a greater frequency of lymph node positivity at presentation. Consequently, significantly lower numbers of male breast cancers were stage I at the time of diagnosis in comparison to female breast cancers. Our results also confirmed the high rate of hormone receptor positivity in male breast cancers (97% ER+, 90% PR+). Unequivocal HER2 positivity rate was 10%.
One of the main goals of this study was to define the IHC profile of male breast cancers for the commonly used diagnostic markers. In this study, we have evaluated male breast cancers against a panel of immunohistochemical stains commonly used to evaluate both breast cancers in general and a tumor of unknown primary in men. In addition to hormone receptors, markers commonly used to confirm breast origin include CK7, GATA3, GCDFP-15, and MGB. CK7 is generally positive in breast, and indeed, 92% of the cases of female breast cancer have been reported to be positive.21 In this cohort of male breast cancers, however, there was a significantly lower level of expression (72% positive) compared with the preexisting data set of female breast cancers. All CK7– cases, however, were positive for ER and GATA3, providing further support for use of a panel of stains in the evaluation of malignancies of uncertain origin that have male breast cancer in the differential diagnosis. Levels of GATA3 expression in male breast cancers have been previously reported to be significantly lower than in female breast cancers.24 However, we found the positivity rate to be high (98% positive) and similar to female breast cancers. GCDFP-15 and MGB expression in male breast cancer in the literature is approximately 55%.14,25 In the current study, we show comparable levels of MGB expression (55%) but higher levels of GCDFP-15 expression (75%). There were 41.8% (23/55) cases positive for both, 14.5% (8/55) negative for both, 30.9% (17/55) positive for GCDFP-15 and negative for MGB, and 10.9% (6/55) positive for MGB but negative for GCDFP-15, supporting that these markers are complementary, as previously suggested, and should both be included in the panel. The high level of AR expression in male breast cancer is well established, a feature seen in this cohort.14,25,26 In males, IHC for AR is commonly performed to establish a diagnosis of prostate cancer, in conjunction with other stains, such as PSA. Expression of AR was high in this cohort (95%) and similar to expression levels reported in female breast cancers. However, none of the male breast cancer cases showed expression for PSA. Although PSA expression in male breast cancer has been previously reported,26 the results of our study suggest that one should consider a new prostatic primary when a tumor at a distant site shows coexpression of AR and PSA. This scenario is not uncommonly identified in male breast cancer cases since these individuals are predisposed to develop both breast and prostatic primary tumors. We suggest using a panel of breast cancer–specific markers (GATA3, GCDFP-15, and MGB) and hormone receptors (ER and PR) to confirm breast as the site of origin. Another interesting finding worth mentioning is reactivity for WT1 in 15% of male breast cancers. It is not entirely surprising since WT1 reactivity has been described in female breast cancers, particularly the mucinous breast tumors.27,28 WT1 is often used as a mesothelial and Müllerian marker (for serous carcinomas) in diagnostic pathology.29 Although staining in the latter is irrelevant for males, WT1 reactivity in pleural or peritoneal fluid may create confusion. However, the H-scores for WT1 in male breast cancers in the current study were generally low, which will help distinguish the tumor cells from generally strong reactivity in mesothelial cells. Nevertheless, we recommend using a panel of epithelial and mesothelial markers (such as BER-EP4, MOC31, WT1, and calretinin) for this distinction.
It is important to note that none of the male breast cancers in this study were positive for lower gastrointestinal tract markers (CK20, CDX2), lung markers (Napsin A, TTF1), renal marker (PAX8), prostatic marker (PSA), or bladder marker (UPII).
In addition to staining for diagnostic markers, we evaluated the diagnostic markers for any association with clinicopathologic factors. Of note, expression of CK7, DPC4, and GCDFP-15 was associated with smaller tumor size. CK7 and DPC4 expression was also associated with lower Nottingham score. Apocrine differentiation in tumors was associated with GCDFP-15 expression, while expression of PR was associated with absence of apocrine differentiation. PR expression was associated with advanced patient age. A total of 28 different diagnostic marker profiles were identified for the four surrogate "molecular" subtypes, demonstrating the inherent immunohistologic heterogeneity of the disease.
Finally, we also analyzed recurrence and survival in this cohort of 61 male breast cancers. Although multivariate analysis could not be performed due to small number of cases, detailed evaluation of the cohort provided unique and useful information. Our results show that the same clinicopathologic factors (stage; receptor status; NPI, which includes tumor size; nodal status; and grade) that are associated with female breast cancer prognosis are also prognostic for male breast cancers. Another interesting observation was regarding the overall benefit of radiation. Since most male breast cancers are treated with mastectomy, many patients do not receive postsurgery radiation. Although radiation had no impact on OS in our cohort, radiation treatment was associated with improved DFS and DDFS. Currently, guidelines for the use of radiation therapy in male breast cancers are similar to its use in female breast cancers. However, given these results and prior published SEER data,23 we believe the role of radiation therapy in the treatment of male breast cancers should be reevaluated. With an average follow-up of 59 months, the OS in our cohort was 70%, but the BCSS was 92%. In the literature, OS for male breast cancer has been variable,10,22,23,30‐34 which likely depends on epidemiologic factors as well as follow-up time. The BCSS in this study is very similar to reported 5-year female BCSS. The difference in OS and BCSS in this study is explained by the high incidence and mortality due to second malignancies in male patients with breast cancer. These findings are somewhat similar to a few recent reports with secondary malignancies occurring in up to 18% of the patients with breast cancer.31,35 Our cohort showed second malignancies in 33% of patients. The varied type of second malignancies in this cohort suggests the possibility of other germline mutations in addition to BRCA2 in male patients with breast cancer. Future studies could include additional genetic testing to further delineate unique mutations in this cohort.
Conclusion
In conclusion, male breast cancers show many similarities to female breast cancers, but subtle differences exist. The patients are slightly older at presentation and are less likely to present at stage I. The tumors are also slightly more proliferative than female counterparts. However, this does not affect BCSS likely due to the effectiveness of the available breast cancer systemic therapies. Nevertheless, OS in male patients with breast cancer could be affected by second malignancies, and these patients should be kept under close clinical surveillance. The immunohistochemical profile reported in this study provides a baseline for comparison in future studies.
Acknowledgment: We thank Amber Luketich of Magee-Womens Hospital for performing immunohistochemistry.