-
PDF
- Split View
-
Views
-
Cite
Cite
Rafal Rak, Andrew Rowley, William Black, Sophia Ananiadou, Argo: an integrative, interactive, text mining-based workbench supporting curation, Database, Volume 2012, 2012, bas010, https://doi.org/10.1093/database/bas010
Close - Share Icon Share
Abstract
Curation of biomedical literature is often supported by the automatic analysis of textual content that generally involves a sequence of individual processing components. Text mining (TM) has been used to enhance the process of manual biocuration, but has been focused on specific databases and tasks rather than an environment integrating TM tools into the curation pipeline, catering for a variety of tasks, types of information and applications. Processing components usually come from different sources and often lack interoperability. The well established Unstructured Information Management Architecture is a framework that addresses interoperability by defining common data structures and interfaces. However, most of the efforts are targeted towards software developers and are not suitable for curators, or are otherwise inconvenient to use on a higher level of abstraction. To overcome these issues we introduce Argo, an interoperable, integrative, interactive and collaborative system for text analysis with a convenient graphic user interface to ease the development of processing workflows and boost productivity in labour-intensive manual curation. Robust, scalable text analytics follow a modular approach, adopting component modules for distinct levels of text analysis. The user interface is available entirely through a web browser that saves the user from going through often complicated and platform-dependent installation procedures. Argo comes with a predefined set of processing components commonly used in text analysis, while giving the users the ability to deposit their own components. The system accommodates various areas and levels of user expertise, from TM and computational linguistics to ontology-based curation. One of the key functionalities of Argo is its ability to seamlessly incorporate user-interactive components, such as manual annotation editors, into otherwise completely automatic pipelines. As a use case, we demonstrate the functionality of an in-built manual annotation editor that is well suited for in-text corpus annotation tasks.
Database URL:http://www.nactem.ac.uk/Argo
Introduction
Text mining (TM) is used increasingly to support biomedical knowledge discovery (1–3), hypothesis generation (4) and to manage the mass of biological literature (5). Its primary goal is to extract new information such as named entities, relations hidden in text and to enable scientists to systematically and efficiently discover, collect, interpret and curate knowledge required for research. Due to the increasing number of articles published each day, the curation of biomedical literature requires the support of automatic tools to retrieve relevant documents and to ease the arduous task of curation (6). TM tools are generally composed of multiple independent processing components bridged together in a pipeline/workflow (7). For instance, before a textual fact about two interacting proteins can be deposited in a database, the processing of the source text would usually involve sentence splitting, tokenization, part-of-speech tagging, protein name recognition and identification and protein relationship extraction. Unfortunately, TM components (resources, tools) available for biomedical TM usually come from different sources and lack interoperability. To overcome the obstacle of combining TM components effectively, the Unstructured Information Management Architecture (UIMA) (8) has been widely adopted. UIMA defines data representations and interfaces to support interoperability between such processing components. UIMA is general enough to handle various kinds of media such as text, audio, image or video; however, in this work we are interested in utilizing a UIMA-based workbench in text processing.
UIMA has drawn significant attention from developers in the biomedical TM community, which has resulted in the development of several resources compliant with the framework, such as Carnegie Mellon University's UIMA Component Repository (http://uima.lti.cs.cmu.edu:8080/UCR/Welcome.do), BioNLP UIMA Component Repository (9), JULIE Lab's UIMA Component Repository (JCoRe) (10) and Open Health NLP (https://cabig-kc.nci.nih.gov/Vocab/KC/index.php/OHNLP). However, UIMA component repositories usually provide only limited support for building task-oriented systems such as a curation system. U-Compare (11,12) addresses this problem by providing a common type system that provides a drag-and-drop graphic user interface (GUI) as well as comparison, evaluation and result visualization mechanisms. U-Compare has a number of useful plug-ins for creating workflows (11) and currently contains more than 50 biomedical TM tools whose performance can be compared within the workbench itself.
Inspired by U-Compare, we have developed Argo—a workbench with a GUI for creating automatic as well as manual annotations derived from TM components. Unlike previous solutions that were almost completely inaccessible to a non-technical audience such as annotators, or database curators, Argo is user- and task oriented, thus suitable for curation teams.
The key features of Argo and how it adds value to U-Compare include:
The user interface is accessible entirely through a web browser. There is no software installation involved.
The processing of user-defined workflows is performed on one or multiple ‘remote’ machines. The user's machine is not used for processing.
The system accommodates users with various areas of expertise. The components available to, e.g., curators are on a higher level of abstraction than those available to text miners who deal with the minutiae of linguistic processing.
Argo incorporates user-interactive processing components designed specifically for a non-technical audience.
Due to its distributed nature, the system supports user collaboration. The users can share their workflows, data and results with others.
Application functionality updates are carried out without the users' involvement.
Argo allows software developers to build their own clients and communicate with the provided web services.
Argo also naturally supports software developers by taking away the burden of having to build peripheral, yet crucial elements of a complete UIMA system, allowing the developers to focus on building individual processing components.
Section 3 describes the above-mentioned features in detail, whereas Section presents generic and real-world use cases.
Related work
TM has been used to enhance the process of manual biocuration before. In the comparative toxicogenomics database (CTD), TM has been used to improve the process of biocuration, but was mostly focused on information retrieval (13). Kleio (14), a TM-based search platform, which is based on semantic types, provides a wide range of semantic search functions that allow users to customize their queries using semantically based facets. Document retrieval based on semantic types radically reduces the search space and reduces false positives. Other uses of TM in biocuration include protein–protein interactions (15) with an estimated 70% reduction in curation workload of yeast–protein interactions using PreBIND/Textomy. Karamanis et al. (6) reported that FlyBase records were completed 20% faster when using PaperBrowser TM tools and Van Auken et al. (16) deployed Textpresso in the curation pipeline of proteins with Gene ontology, reporting an efficiency increase of 8-fold over manual curation.
Most of the reported TM tools for curation are used for specific databases and tasks. For instance, ODIN (17), a web-based tool for the curation of biomedical literature, is equipped with a fixed set of biomedical named entity recognizers and comes with a user interface, which is tailored for tasks related to the recognized named entities. Argo, on the other hand, is not related to any specific task. To the contrary, it allows the users to define their own tasks. Therefore, Argo is not only a collaborative and modular system of information extraction that links the manual and automated annotation of textual documents, but it also allows the curators to build workflows (processing pipelines), which define the task at hand.
The idea of providing a convenient interface for users to build processing pipelines has been proposed previously. Notable examples of such systems include Taverna (18), Galaxy (19) and GATE (General Architecture for Text Engineering) (20). As far as systems designed specifically to handle text processing are concerned—apart from already discussed U-Compare—GATE is the most closely related system. The key difference between GATE and Argo (or any other UIMA-based system) is that GATE is primarily intended to support programmers by providing an integrated development environment (IDE), rather than being a workflow-supporting framework. Moreover, GATE does not define any data type hierarchy capable of assisting interoperability between processing components. However, the most pronounced advantage of Argo over other systems lies in its ability to seamlessly incorporate user-interactive components into otherwise completely automatic pipelines as well as its ability to accommodate users with various areas of expertise. To the best of our knowledge, Argo is the first such solution.
One of the user-interactive components, the annotation editor, was designed based on experiences gained while using XConc (http://www-tsujii.is.s.u-tokyo.ac.jp/GENIA/home/wiki.cgi?page=XConc+Suite). XConc is an annotation suite that was developed as a plug-in for Eclipse, an IDE designed primarily for software developers. Among other tasks, XConc was used for mapping text with manually curated pathways (21). Although it allows for rich expressive power, its dependency on Eclipse makes its usage challenging for a non-technical audience. Argo, in contrast, uses a web browser, which is a more intuitive, general-audience medium.
System overview
User interface
The user interface of Argo is accessible entirely through a web browser. The browser communicates with the server through a series of asynchronous calls (i.e. there is no navigating between or reloading pages), which makes user interaction with the system fast and non-distracting.
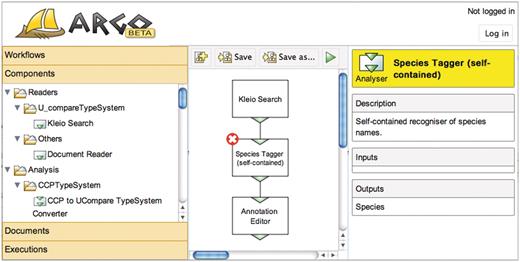
A screen capture of the interface in use is shown in Figure 1. The main panel of the interface is a canvas onto which the users drop graphical elements representing, in most cases, processing components from the side panel. The side panel is the central navigation element organized into categories of objects that the users are allowed to manipulate. These are:
Documents
Documents are resources containing text and are the primary subject of processing. Documents may also be the result of processing, e.g. a workflow may end with a consumer that saves the extracted annotations into an XML file (which can be further reused in other workflows or exported to a relational database). Multiple documents may be grouped in folders that in turn can be further nested and ultimately resemble the familiar filesystem.
Components
The processing components form the most intrinsic objects of the system from the user's perspective. Processing components are enclosed algorithms that, in their most typical use, process the input data and produce its annotation-augmented version as an output. Each processing component defines the input and output types it is capable of handling. Thus, a single processing component expects an input CAS to contain annotations of particular types. A component may define multiple input and output types.
Workflows
Multiple processing components interconnected in a specific, user-defined order form a workflow. Workflows are created and manipulated by selecting the processing components from the side panel and placing them onto the diagramming canvas. Workflows are the primary subject of ‘executions’.
Executions
This category lists the current and past executions of workflows. It consists of information such as the time of execution, duration and current progress. An execution does not terminate unless it is complete or the user explicitly deletes it. The user can start running the execution, close the browser and then come back to it later to see the current progress of execution. This is a useful feature particularly with computationally expensive workflows or large inputs.
Processing components and workflows
UIMA differentiates between two types of processing components, namely ‘readers’ and ‘analysis engines’. Whereas the former act as source components that read the input data to be processed further in the pipeline, the latter update CASes with new annotations and pass the updated versions further. ‘Consumers’ constitute a notable subclass of analysis engines. They are capable of serializing CASes to storable formats, such as plain-text files, XML files, databases, etc.
The system comes with a predefined set of processing components and workflows for various tasks, from sentence splitting and tokenization, to named-entity recognition, to database storage. Argo also allows the users to deposit their own components as long as they comply with the UIMA specification.
A special type of processing components is a user-interactive component that requires input from the user. If user-interactive components are present in a workflow, the processing of the workflow pauses at which point the user is expected to provide some sort of input, which in most cases will be manual annotation. Argo provides an example of a user-interactive component: a general-use annotation editor. The editor allows for adding new span-of-text annotations, removing or modifying previously identified annotations and adding metadata. The annotated spans of text can embed or even overlap with other spans. In both cases the editor marks and displays the annotations in a lucid and visually unambiguous manner. Figure 2 shows a screen capture of the annotation editor in action.
Annotation editor in action: the user is about to add an annotation manually to automatically pre-annotated text.
A read-only version of the annotation editor is used to visualize annotations in Argo. This is a convenient way of quickly verifying the annotations produced with the current workflow before serializing the results to other—more useful—formats.
Technology
The user interface brings together technologies such as Google Web Toolkit (GWT) (http://code.google.com/webtoolkit), a development toolkit for building web-based applications, and Scalable Vector Graphics (SVG) (http://www.w3.org/TR/SVG/), an open standard for describing vector graphics, which is widely supported by web browsers and is heavily utilized in the annotation editor.
The client–server communication is accomplished through well-established web service protocols, SOAP (http://www.w3.org/TR/soap12-part1) and REST (22). The inclusion of web-service protocols is a purposeful effort to allow software developers who wish to build their own systems to connect directly to the Argo server (in fact, a number of dedicated load-balanced servers). For instance, a workflow created with the Argo interface, may be used directly in the user's client. Additionally, the distributed nature of the system means that the custom-built clients will be able to immediately take advantage of any changes made by workflow designers, which abstracts away the inner workings of the custom-built client from the workflow and its future modifications. This significantly accelerates the collaborative development cycle.
Use cases
Workflows
The following generic use case is based on a simple task of annotating the occurrences of species names in a stream of documents coming from the output of a search engine. One of the requirements is that the task should be carried out in a semi-automatic fashion, where the automatic part is responsible for tagging the species names to the best of its capacity, whereas the manual part involves verifying and possibly editing the automatically recognized instances. The use case illustrates two scenarios of using the workbench by two types of users: one without a technical background and another with a certain level of proficiency in natural language processing (NLP).
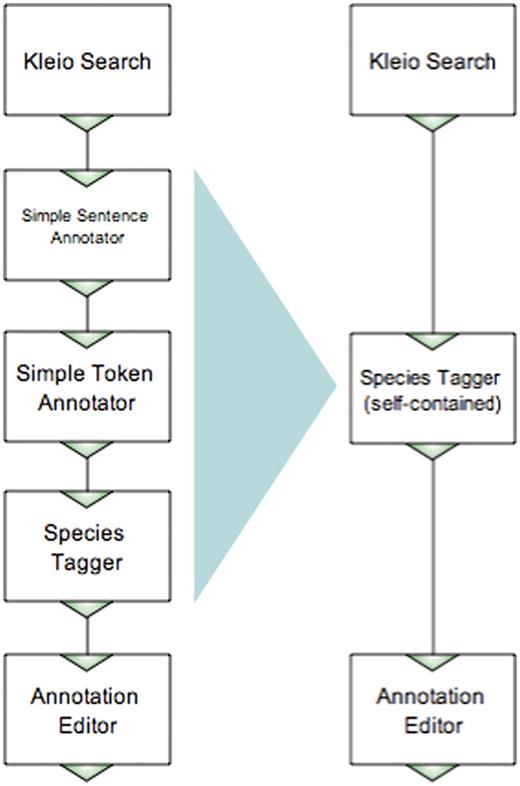
Figure 3 shows two workflows, one built by the user with an NLP background, and another by the non-technical user. The workflows include two components common for both: the Kleio search that retrieves MEDLINE abstracts matching a query specified as the component's parameter, and the Argo manual annotation editor. The user with an NLP background would most likely build the species names recognizer (tagger) using atomic NLP components such as a sentence annotator, a tokenizer and a dictionary-based tagger, as shown on the left-hand side of Figure 3. However, the non-technical user, who is not familiar with NLP, would possibly want to use a self-contained species tagger—one that contains all the necessary NLP processing inside—as shown on the right-hand side of Figure 3. Fortunately, Argo supports ‘aggregate components’, i.e. components that contain other (atomic or aggregate) components. Thus, the specialist can take care of the minutiae of NLP processing and create an aggregate component (in this case consisting of the sentence annotator, the tokenizer and the tagger), which will later be available to the non-technical user.
Examples of workflows performing the same task, built in Argo by an NLP expert and a non-technical user.
It is worth noting that, although the workflows depicted in the figure are complete (in the sense that they are ready to be executed), they lack an end component that would write the processed data to a permanent format such as database. In most curation tasks, this component needs to be implemented and tailored to meet the requirements of the underlying task- and/or organization-specific database structure.
Annotation editor
The Argo annotation editor is currently being used in an annotation task, whose goal is to extract interactions between enzymes and marine drugs in over 230 full-text articles from biomedical journals. Two annotators work individually and are supported by processing components, which automatically find the named entities in text and identify phrases with potential mentions of interactions. The automatically recognized entities consist of enzymes and marine drugs. The annotators’ task is to verify if the automatically identified named entities are correct, and manually annotate words or phrases signalling interactions between the named entities, as well as specify the roles, such as ‘agent’ and ‘target’, played by each of the named entities participating in the interaction. Figure 1 is, in fact, a screen capture of this task's setup.
The annotators have a background in biology and prior experience in using annotation tools, such as XConc. For this task, they are asked to provide ‘complete’ annotation, i.e. to create a ‘gold-standard corpus’. This corpus will later be used to train automatic processing components, which, in turn, will be utilized to find enzyme–drug interactions on a broader scale.
Statistics derived from a subset of 30 fully annotated articles show that on average there are about 128 automatically recognized entities (enzymes, drugs and phrases) per document, of which the annotators removed (as being false positive) on average about 4.6, modified (e.g. extended or narrowed down the annotation's boundaries) about 1.7 and added a further 6 per document. Table 1 shows a more detailed view.
Average number of annotations per document per label for the enzyme–drug interactions task
| Label . | Pre-annotated . | Added, N (%) . | Modified, N (%) . | Removed, N (%) . |
|---|---|---|---|---|
| Enzyme | 114.3 | 4 (3.5) | 0.7 (0.6) | 2.7 (2.3) |
| Drug | 10.5 | 1 (9.9) | 0.5 (4.6) | 0.5 (4.8) |
| Phrase | 3.6 | 1 (27.8) | 0.5 (13.4) | 1.5 (40.7) |
| Total | 128.4 | 6 (4.7) | 1.7 (1.3) | 4.6 (3.6) |
| Label . | Pre-annotated . | Added, N (%) . | Modified, N (%) . | Removed, N (%) . |
|---|---|---|---|---|
| Enzyme | 114.3 | 4 (3.5) | 0.7 (0.6) | 2.7 (2.3) |
| Drug | 10.5 | 1 (9.9) | 0.5 (4.6) | 0.5 (4.8) |
| Phrase | 3.6 | 1 (27.8) | 0.5 (13.4) | 1.5 (40.7) |
| Total | 128.4 | 6 (4.7) | 1.7 (1.3) | 4.6 (3.6) |
Average number of annotations per document per label for the enzyme–drug interactions task
| Label . | Pre-annotated . | Added, N (%) . | Modified, N (%) . | Removed, N (%) . |
|---|---|---|---|---|
| Enzyme | 114.3 | 4 (3.5) | 0.7 (0.6) | 2.7 (2.3) |
| Drug | 10.5 | 1 (9.9) | 0.5 (4.6) | 0.5 (4.8) |
| Phrase | 3.6 | 1 (27.8) | 0.5 (13.4) | 1.5 (40.7) |
| Total | 128.4 | 6 (4.7) | 1.7 (1.3) | 4.6 (3.6) |
| Label . | Pre-annotated . | Added, N (%) . | Modified, N (%) . | Removed, N (%) . |
|---|---|---|---|---|
| Enzyme | 114.3 | 4 (3.5) | 0.7 (0.6) | 2.7 (2.3) |
| Drug | 10.5 | 1 (9.9) | 0.5 (4.6) | 0.5 (4.8) |
| Phrase | 3.6 | 1 (27.8) | 0.5 (13.4) | 1.5 (40.7) |
| Total | 128.4 | 6 (4.7) | 1.7 (1.3) | 4.6 (3.6) |
Together with the additional annotations with labels that had not been automatically pre-annotated, the number of annotation actions (additions, modifications and removals) came to about 24 on average per document, which constitutes <16% of the total number of extracted annotations, i.e. the automatic processing was responsible for the completion of ∼84% of the task.
The inter-annotation agreement on the interaction mentions (the main objective of the task) was at the level of 55% in F1 score. However, relaxed matching shows that the agreement was actually as high as 82%. The relaxed matching takes into consideration overlapping annotations, and not only exact matches. For instance, one of the annotators would consistently add modality (e.g. ‘would affect’ instead of simply ‘affect’) or manner (e.g. ‘significantly increase’ instead of ‘increase’). These inconsistencies between annotators come from the annotators’ interpretation of the task-specific annotation guidelines, and are not the result of inability to operate the annotation editor.
Apart from being given the annotation guidelines with a few examples with screen captures of the system, the annotators did not receive any training on how to use the annotation editor. Both annotators agreed that the editor is intuitive, as well as easier and faster to use than the tools they had previously used (e.g. XConc). They spent 12.6 min on average per full-text article. Given that the average size of the articles was about 4800 words, this results in about 380 words/min, which is slightly higher than the average reading speed of an average reader. This may stem from the fact that the annotators were not required to thoroughly understand the article and most likely elected to skim sections that did not mention enzymes or drugs.
In comparison to XConc, the Argo annotation editor's biggest productivity enhancement is a sharp learning curve. XConc requires familiarity with Eclipse (an IDE it is embedded in), and thus its usage might be prohibitive to the non-technical user. The Argo editor, on the other hand, does not require any prior training and its usage shares similarities with general-purpose tools, such as word processors.
Ongoing and future work
Ongoing work includes improving the system access management that is a key feature in achieving true distributed user collaboration. Users will be able to collaborate in groups and share—individually or as a whole—anything from the input documents, to processing components, to workflows, to the partial or full results of workflow executions.
One of the most significant advantages of Argo over other systems will come with the addition of an ‘annotation task editor’. The editor will allow the user to define flexible type systems with sophisticated relationships between the types, ‘type constraints’, a novel approach to UIMA type systems. The user will be capable of expressing constraints such as ‘type Binding must be associated with at least two Protein types via the Theme roles’. The editor will provide a GUI with the familiar drag-and-drop mechanism that will make the definition of the annotation task more intuitive and unambiguous (which is often a problem with annotation guidelines written in plain language). Once an annotation task is defined, it will serve as an annotation guideline for manual annotation (after automatic and unambiguous translation to the plain-language version), a validator of manual and automatic processing components, as well as a configuration component for automated recognition tools.
We also continue to introduce new components to the ever-increasing library of processing components. In particular, we are planning on adding two Conditional Random Field-based (23) generic annotation components: one for building a statistical model representing already annotated data, and another for annotating new data based on the previously built model that will be passed to the latter as a parameter. The two components will particularly be of interest to users with no or limited technical background who have a sample of manually annotated data and wish to increase it by means of automatic processing.
Conclusions
By using an interoperable framework wrapped in an intuitive user interface, Argo is able to leverage existing and established TM tools using a modular approach. This enables user- and task-oriented applications. The modular approach allows the system to be adapted and its parts reused for new domains, and for analysis and comparison pipelines using different permutations of modules to be evaluated, producing incremental performance gains on established tasks. As we are leveraging NaCTeM's U-Compare platform and TM workflow environment, we can integrate them seamlessly into the curation pipeline linking them with the manual and automated annotation of textual documents.
Funding
Biotechnology and Biological Sciences Research Council (BB/G013160/1 Automated Biological Event Extraction from the Literature for Drug Discovery); National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (HHSN272200900040C to Bruno Sobral); National Centre for Text Mining is funded by Joint Information Systems Committee (JISC). Funding for open access charge: Biotechnology and Biological Sciences Research Council (BB/G013160/1 Automated Biological Event Extraction from the Literature for Drug Discovery).
Conflict of interest. None declared.
Acknowledgements
The authors would like to specially thank BalaKrishna Kolluru and Riza Theresa Batista-Navarro who provided access to the enzyme–drug annotation task they supervise, which was used to demonstrate some of the capabilities of the system in this work.