-
PDF
- Split View
-
Views
-
Cite
Cite
Catalina O. Tudor, Cecilia N. Arighi, Qinghua Wang, Cathy H. Wu, K. Vijay-Shanker, The eFIP system for text mining of protein interaction networks of phosphorylated proteins, Database, Volume 2012, 2012, bas044, https://doi.org/10.1093/database/bas044
Close - Share Icon Share
Abstract
Protein phosphorylation is a central regulatory mechanism in signal transduction involved in most biological processes. Phosphorylation of a protein may lead to activation or repression of its activity, alternative subcellular location and interaction with different binding partners. Extracting this type of information from scientific literature is critical for connecting phosphorylated proteins with kinases and interaction partners, along with their functional outcomes, for knowledge discovery from phosphorylation protein networks. We have developed the Extracting Functional Impact of Phosphorylation (eFIP) text mining system, which combines several natural language processing techniques to find relevant abstracts mentioning phosphorylation of a given protein together with indications of protein–protein interactions (PPIs) and potential evidences for impact of phosphorylation on the PPIs. eFIP integrates our previously developed tools, Extracting Gene Related ABstracts (eGRAB) for document retrieval and name disambiguation, Rule-based LIterature Mining System (RLIMS-P) for Protein Phosphorylation for extraction of phosphorylation information, a PPI module to detect PPIs involving phosphorylated proteins and an impact module for relation extraction. The text mining system has been integrated into the curation workflow of the Protein Ontology (PRO) to capture knowledge about phosphorylated proteins. The eFIP web interface accepts gene/protein names or identifiers, or PubMed identifiers as input, and displays results as a ranked list of abstracts with sentence evidence and summary table, which can be exported in a spreadsheet upon result validation. As a participant in the BioCreative-2012 Interactive Text Mining track, the performance of eFIP was evaluated on document retrieval (F-measures of 78–100%), sentence-level information extraction (F-measures of 70–80%) and document ranking (normalized discounted cumulative gain measures of 93–100% and mean average precision of 0.86). The utility and usability of the eFIP web interface were also evaluated during the BioCreative Workshop. The use of the eFIP interface provided a significant speed-up (∼2.5-fold) for time to completion of the curation task. Additionally, eFIP significantly simplifies the task of finding relevant articles on PPI involving phosphorylated forms of a given protein.
Database URL:http://proteininformationresource.org/pirwww/iprolink/eFIP.shtml
Introduction
Post-translational modifications play a fundamental role in regulating the activity, location and function of a wide range of proteins. In particular, protein phosphorylation by protein kinases and dephosphorylation by phosphatases play a major role in almost all critical cellular events, such as cell metabolism regulation, cell division, cell growth and differentiation. Often, protein phosphorylation results in some functional impact. For instance, proteins can be phosphorylated on different residues, leading to either activation or down-regulation of their activities, alternative subcellular locations and/or interaction with distinct binding partners.
As an example, protein Smad2 has different phosphorylation sites leading to various phosphorylation states which determine its interaction partners, subcellular location and activity, as illustrated in the example sentence below. The phosphorylation and the interaction mentions are emphasized in bold, and the impact of phosphorylation on the interaction in italics.
TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required forSmad2-Smad4 complex formation and signaling. (PMID 9346908)
We have developed Extracting Functional Impact of Phosphorylation (eFIP) (1) to extract such information from PubMed abstracts. The first step is to detect mentions of phosphorylation and protein–protein interaction (PPI) involving the phosphorylated protein. Once the phosphorylation and interaction mentions are detected, the second step is to identify a possible relation between the two events. We look for temporal relations and subsequent causal relations, as these are important biological context of phosphorylation. The types of PPIs captured by eFIP include interactions between a protein and another protein, a protein complex, a protein region or a protein class.
Protein interaction data involving phosphorylated proteins are not yet well represented in public databases. However, this information is critical for the understanding of protein networks and prediction of the functional outcomes. Information about phosphorylated proteins extracted by eFIP from the scientific literature and validated by curators is captured in the Protein Ontology (PRO) (2). PRO provides an ontological structure to capture knowledge about protein classes, multiple protein forms (e.g. isoforms and post-translationally modified forms) and protein complexes in the Open Biological and Biomedical Ontologies Foundry framework, thereby allowing precise definition of protein objects in biological context and specification of relationships that describe properties of those entities.
The main contributions of this article include: (i) the development of a system for detecting involvement of phosphorylated proteins in protein interactions, (ii) the design of syntactic patterns and lexical features to detect possible impact of phosphorylation on interaction, (iii) the manual curation of a set of abstracts as the gold standard literature corpus, which is made publically available and (iv) the evaluation of the system in terms of accuracy and usefulness for biocuration.
The rest of the article is organized as follows. The Related Work section briefly describes the various work related to the different components of eFIP. The system itself, each individual component and the web interface are described in detail in the Materials and Methods section. We describe the evaluations conducted on eFIP and analyze the results and the feedback obtained from biocurators who participated in the BioCreative-2012 Workshop in the Results and Discussion sections. We conclude and describe future work at the end of the manuscript.
Related work
For document retrieval relevant to a given protein, we use Extracting Gene Related ABstracts (eGRAB) (3) (described in the Materials and Methods section). A critical step in eGRAB is the disambiguation of gene names. It has been noted that gene names are highly ambiguous, not only across species but also within the same species, as well as with regard to other biomedical abbreviations (4,5). Other approaches have been suggested in the biomedical literature for disambiguating gene names (6,7) and biomedical abbreviations in general (8,9). However, eGRAB was the only system that fit our requirements of finding and retrieving all documents relevant to a given protein from the Medline database. We were able to modify its output to match eFIP’s requirements.
For the detection of phosphorylation mentions in text, we integrated Rule-based LIterature Mining System for Protein Phosphorylation (RLIMS-P) (10,11) (see Materials and Methods section) in our pipeline. MinePhos (12) is another system that extracts phosphorylation information from the literature. Phosphorylation events were also sought, among other events, by the BioNLP 2011 Shared Task (13), although the task did not cover kinase extraction. Comparing with other systems, RLIMS-P provides a broader coverage of kinds of phosphorylation mentions in texts. RLIMS-P also continues to be improved—using a large set of rules coupled with machine learning and other Natural language processing (NLP) techniques to detect complex phosphorylation mentions across texts, thus, is particularly appropriate to use in our pipeline.
Several different approaches have been suggested for the detection of PPIs. The AkaneRE system (14) uses machine learning and syntactic features to detect PPIs, among other types of relation extractions. Protein Interaction information Extraction system (PIE) (15) utilizes natural language processing techniques and machine learning methodologies that do not rely on syntactic features to predict PPI sentences. Xiao et al. (16) applied maximum entropy machine learning method to extract PPI information from the literature. Our decision to build an in-house PPI system is because the PPI systems described in the literature are either not available for download or not easily adaptable to our needs (for more details, see the description of the PPI module within the Materials and Methods section).
The impact module of eFIP detects temporal and causal relations between phosphorylation and interaction events in a sentence. There have been many approaches on detection of temporal relations in a sentence. Lapata and Lascarides (17) employed linguistic features similar to the ones used for eFIP (such as verb classes and argument relations) to learn temporal relations. Mani et al. (18) worked on events spanning multiple sentences for temporally ordering and anchoring events in natural language text. Girju (19) devised a classification of causal questions and tested the procedure on a question answering system. Blanco et al. (20) presented a supervised method for the detection and extraction of causal relations from open domain text. Although much effort has been put into detection of events in biomedical literature, we note the highly relevant work of Raghavan et al. (21) for temporal classification of medical events, the work of Miwa et al. (22) for event extraction with complex event classification and the short paper by van der Horn et al. (23) for detection of causal relations. The BioNLP 2011 Shared Tasks focused on regulation of events, similar to causality relations, but did not address temporal relations. As eFIP focuses on a particular type of temporal and causal relation between a phosphorylation event and a PPI event involving the same phosphorylated protein, the more general approaches mentioned earlier could not be used without considering additional rules specific to this case. With the selection of different software components discussed earlier, we have developed a text mining system specially tailored for mining of protein interaction networks of phosphorylated proteins. Specifically, the eFIP system pipeline (see Materials and Methods section) finds relevant abstracts mentioning phosphorylation and PPI along with its potential impact—a unique functionality not provided by any other system to the best of our knowledge.
Materials and methods
The overview of the eFIP system pipeline is illustrated in Figure 1. The system involves a document retrieval module (eGRAB) to gather all documents mentioning a specific protein, an extraction component (RLIMS-P) to identify mentions of phosphorylation, a PPI module to detect PPIs of phosphorylated proteins and an impact module to identify temporal and causal relations between phosphorylation and interaction events involving the same protein, where applicable. Because a majority of our components identify information based on matching lexico-syntactic patterns, the text is first tagged with part-of-speech types for each word, and subsequently chunked in noun phrases (NPs) and verb groups (VGs). We use the GENIA part-of-speech tagger (24) and a chunker/shallow parser that we have trained on the GENIA corpus.
The eFIP text mining system overview. The pipeline consists of four components to process: (1) retrieval of all documents relevant to a given protein (eGRAB), (2) extraction of phosphorylation mentions (kinase, substrate and site) in these documents (RLIMS-P), (3) extraction of PPI mentions (protein interactants and type of interaction) (PPI module) and (4) detection of phosphorylation-interaction relations (impact module).
Possible inputs are a gene/protein name or identifier, or a list of PubMed identifiers (PMIDs). Although a gene/protein identifier is species-specific, we do not limit the abstracts to the ones mentioning the gene/protein with the corresponding species. Instead, we use the identifier to retrieve all possible names of the gene/protein, which are then used as input in the document retrieval module. Currently, eFIP considers only the Abstract section of a PubMed paper. Extension of eFIP to the Results and Discussion sections of a paper is in development.
eGRAB
eGRAB is used to gather the literature for a given gene/protein. eGRAB starts by gathering all possible names and synonyms of a gene/protein from EntrezGene and UniProtKB. Variations in names are considered to accommodate inclusion/exclusion of hyphenation and spacing (e.g. Bmp2, Bmp-2 and Bmp 2). In an effort to limit the retrieval of too many irrelevant documents, synonyms of a gene/protein are restricted to the ones found in entries for related organisms. The names, synonyms and variations of these are then used as an expanded query to retrieve Medline abstracts. For the retrieval of abstracts, we used the Lucene search engine (25).
Names of genes/proteins are often ambiguous. For instance, ‘CAD’ is not only the official symbol for various different genes (e.g. ‘Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase and dihydroorotase’, ‘Conserved ATPase domain protein’, ‘Caldesmon’ and ‘Caudal’) but also used in Medline with multiple other senses, such as ‘coronary artery disease’, ‘computer-aided design’ and ‘charge aerosol detector’. There are somewhere in the vicinity of 50 different senses for ‘CAD’ in Medline. Therefore, whenever a search is conducted on ‘CAD’, documents mentioning all these senses are retrieved. A PubMed search for ‘CAD’ in the title or abstract retrieved more than 13 000 hits at the time eGRAB was evaluated. However, based on our analysis, only 63 of these mentioned ‘CAD’ in the context of the gene Caudal, for instance.
eGRAB automatically identifies documents that mention an ambiguous name in the context of our interest. eGRAB creates language models for every possible sense of a name, by looking at the words and phrases mentioned together in the context of that sense. For every abstract containing an ambiguous symbol unaccompanied by its expanded form, eGRAB looks at the document’s similarity to the various language models created, and chooses the sense that is closest in similarity. eGRAB is currently being used in other systems. A detailed description of its approach and an evaluation are provided in (3), in the context of eGIFT, a system for the automatic extraction of genic information from text.
RLIMS-P
RLIMS-P is a system designed for extracting protein phosphorylation information from text. It extracts the three objects involved in this process: the protein kinase, the phosphorylated protein (substrate) and the phosphorylation site (residue or position being phosphorylated). An example of information extracted by RLIMS-P is shown highlighted in the following sentence:
[TbetaRI]_kinase [phosphorylation] of [Smad2]_phosphorylated_protein on [Ser465 and Ser467]_site is required for Smad2-Smad4 complex formation and signaling. (PMID 9346908)
RLIMS-P utilizes extraction rules that cover a wide range of patterns, including some specialized terms used only with phosphorylation. Additionally, RLIMS-P employs techniques to combine information found in different sentences, because rarely are the three objects (kinase, substrate and site) found in the same sentence. RLIMS-P has been benchmarked and the results are presented in (10). A detailed description of the system can be found in (11) and the system itself is available for online text mining at: http://proteininformationresource.org/pirwww/iprolink/rlimsp.shtml.
PPI module
The PPI tool was designed to include among others: (i) the ability to detect interactions involving only one partner when the other partner is implicit, (ii) the ability to detect anaphora resolution when one of the partners or both are described by pronouns (e.g. ‘it’, ‘they’ as can be seen in the second and third examples in Table 1) and (iii) the ability to detect termination of an interaction (e.g. ‘dissociates’) or lack of an interaction (e.g. ‘cannot bind’).
Example patterns that capture PPI mentions
| Pattern . | Example phrase capturing the pattern . |
|---|---|
| NP_P1 NP_int Prep_from NP_P2 | … 14-3-3binding and dissociation from Bcl-XL |
| NP_its NP_int Prep_with NP_P2 | … itsassociation with Bcl-XL |
| NP_it VG_int Prep_with NP_P2 | … itdimerizes with Bcl-XL… |
| NP_int Prep_of NP_P1 Prep_to NP_P2 | … the binding of BAD to Bcl-XL |
| NP_P1 VG_int Prep_with NP_P2 | PP2A and 14-3-3 can interact with FOXO1… |
| Pattern . | Example phrase capturing the pattern . |
|---|---|
| NP_P1 NP_int Prep_from NP_P2 | … 14-3-3binding and dissociation from Bcl-XL |
| NP_its NP_int Prep_with NP_P2 | … itsassociation with Bcl-XL |
| NP_it VG_int Prep_with NP_P2 | … itdimerizes with Bcl-XL… |
| NP_int Prep_of NP_P1 Prep_to NP_P2 | … the binding of BAD to Bcl-XL |
| NP_P1 VG_int Prep_with NP_P2 | PP2A and 14-3-3 can interact with FOXO1… |
‘NP’ stands for noun phrase, ‘NP_P’ stands for a noun phrase that holds a protein name and ‘NP_int’ stands for a noun phrase holding a trigger word for interaction (e.g. ‘binds’, ‘binding’, ‘interacts’, ‘interaction’, etc.). ‘VG_int’ stands for a verb group containing a trigger word for interaction. ‘Prep’ stands for preposition, and the actual preposition is given after the underscore line. Pronouns are also allowed as interactant, and we mark them with ‘NP_its’, ‘NP_it’, ‘NP_they’, etc.
Example patterns that capture PPI mentions
| Pattern . | Example phrase capturing the pattern . |
|---|---|
| NP_P1 NP_int Prep_from NP_P2 | … 14-3-3binding and dissociation from Bcl-XL |
| NP_its NP_int Prep_with NP_P2 | … itsassociation with Bcl-XL |
| NP_it VG_int Prep_with NP_P2 | … itdimerizes with Bcl-XL… |
| NP_int Prep_of NP_P1 Prep_to NP_P2 | … the binding of BAD to Bcl-XL |
| NP_P1 VG_int Prep_with NP_P2 | PP2A and 14-3-3 can interact with FOXO1… |
| Pattern . | Example phrase capturing the pattern . |
|---|---|
| NP_P1 NP_int Prep_from NP_P2 | … 14-3-3binding and dissociation from Bcl-XL |
| NP_its NP_int Prep_with NP_P2 | … itsassociation with Bcl-XL |
| NP_it VG_int Prep_with NP_P2 | … itdimerizes with Bcl-XL… |
| NP_int Prep_of NP_P1 Prep_to NP_P2 | … the binding of BAD to Bcl-XL |
| NP_P1 VG_int Prep_with NP_P2 | PP2A and 14-3-3 can interact with FOXO1… |
‘NP’ stands for noun phrase, ‘NP_P’ stands for a noun phrase that holds a protein name and ‘NP_int’ stands for a noun phrase holding a trigger word for interaction (e.g. ‘binds’, ‘binding’, ‘interacts’, ‘interaction’, etc.). ‘VG_int’ stands for a verb group containing a trigger word for interaction. ‘Prep’ stands for preposition, and the actual preposition is given after the underscore line. Pronouns are also allowed as interactant, and we mark them with ‘NP_its’, ‘NP_it’, ‘NP_they’, etc.
The PPI module extracts text fragments (evidence) for each of the parts involved in an interaction: first interactant, second interactant (if available) and the type of PPI (e.g. binding, dissociation and complex). The primary engine of this module is an extensive set of rules specialized to detect patterns of PPI mentions (e.g. patterns for matching a PPI in a sentence are provided in Table 1). The NPs that are detected as the interacting partners are further sent to a gene mention tool (26) to confirm whether they are genuine protein mentions. Additionally, we try to resolve pronouns (e.g. ‘it’, ‘their’, etc.) and relative pronouns (e.g. ‘which’, ‘that’, etc.) to protein names. Consider the following sample phrase:
A variety of survival signals are reported to induce the phosphorylation of BAD at Ser(112) or Ser(136), triggering itsdissociation from Bcl-X(L). (PMID 10880354)
‘it’ is identified as the first argument of the dissociation, which prompts the need to further identify the actual protein (Bad) that gets dissociated from Bcl-X(L).
In eFIP, PPIs include the following types: interactions between proteins, interactions between a protein and a protein complex, interactions between a protein and a protein region and interactions between a protein and a class of proteins.
Impact module
The goal of this module is to find information about the ability of phosphorylated proteins to interact with other proteins. We found that whenever there is a relationship between phosphorylation and interaction events, this relationship is described almost exclusively in the same sentence. Given a sentence that contains mentions of both phosphorylation (as given by the RLIMS-P module) and interaction (as given by the PPI module), the next step is to detect if there is a ‘temporal’ relation between them (in which the phosphorylation is found to occur before the interaction), and if so, whether we can determine a ‘causal’ relationship as well.
The types of causal relations can be ‘positive’ (phosphorylation of A increases its binding to B) or ‘negative’ (phosphorylated A dissociates from B). If no causal relationship can be determined, but a temporal relationship identifies that the phosphorylation happens before the interaction, then we say the relationship between the phosphorylation and the interaction is ‘neutral’ (phosphorylated A binds B).
For example, consider the following sentence:
PhosphorylatedBadbinds to the cytosolic 14-3-3 protein. (PMID 11526496)
In this example, it is clear that the phosphorylation happens before the binding, as one of the interactants, Bad, is reported to be ‘phosphorylated’. However, whether the phosphorylation has any impact on the binding itself (i.e. if 14-3-3 binds to Bad regardless of its form, phosphorylated or non-phosphorylated) is not clearly stated in this sentence. Thus, the phosphorylation-interaction relationship in this example is neutral.
In contrast, the next sentence not only points to a temporal relationship in which phosphorylation happens before the interaction but also describes a causal relation (i.e. how the interaction is a consequence of the phosphorylation):
Badphosphorylation induced by survival factors leads to its preferential binding to 14-3-3 and suppression of the death-inducing function of Bad. (PMID 10579309)
We studied a development set of 300 sentences marked with involvement of phosphorylated proteins in interactions, and designed a set of rules to determine whether a sentence contains the type of information sought for this task. The rules are based on the features described in Table 2, and each rule is assigned a confidence score based on how strong it was in the development set.
Features used in the detection of phosphorylation-interaction relations
| Type . | Feature . | Description . |
|---|---|---|
| T, C | SSI | Substrate is the same as interactant |
| T | IMP | One of the interactants is mentioned as being ‘phosphorylated’ (phosphorylated A binds to B) |
| T | CONJ | P and I are mentioned in a conjunction (there are five types of conjunctions captured in five different features) |
| C | ACTION | P and I are mentioned in a Subject–Verb–Object relationship (A phosphorylation leads to interaction with B) |
| T, C | DEPEND | I mentioned to be dependent on P (phosphorylation-dependent interaction of A to B) |
| T | PFIRST | P mentioned before I in the sentence |
| T | IFIRST | I mentioned before P in the sentence |
| T | WLR | There is a word/phrase between P and I hinting to a directionality of events from left to right (leads to) |
| T | WRL | There is a word/phrase between P and I hinting to a directionality of events from right to left (requires) |
| C | NEG | One of the events or the action is being negated (phosphorylated A does not bind to B) |
| C | HEDGE | One of the events or the action is mentioned with hedging (phosphorylated A might bind to B) |
| T | RELAPPB | P or I is mentioned in a relative clause or appositive referring to a protein (A, which interacts with B, is phosphorylated by C) |
| T | RELAPPG | I is mentioned in a relative clause or appositive referring to the phosphorylation (phosphorylation of A, which increased the interaction with B) |
| Type . | Feature . | Description . |
|---|---|---|
| T, C | SSI | Substrate is the same as interactant |
| T | IMP | One of the interactants is mentioned as being ‘phosphorylated’ (phosphorylated A binds to B) |
| T | CONJ | P and I are mentioned in a conjunction (there are five types of conjunctions captured in five different features) |
| C | ACTION | P and I are mentioned in a Subject–Verb–Object relationship (A phosphorylation leads to interaction with B) |
| T, C | DEPEND | I mentioned to be dependent on P (phosphorylation-dependent interaction of A to B) |
| T | PFIRST | P mentioned before I in the sentence |
| T | IFIRST | I mentioned before P in the sentence |
| T | WLR | There is a word/phrase between P and I hinting to a directionality of events from left to right (leads to) |
| T | WRL | There is a word/phrase between P and I hinting to a directionality of events from right to left (requires) |
| C | NEG | One of the events or the action is being negated (phosphorylated A does not bind to B) |
| C | HEDGE | One of the events or the action is mentioned with hedging (phosphorylated A might bind to B) |
| T | RELAPPB | P or I is mentioned in a relative clause or appositive referring to a protein (A, which interacts with B, is phosphorylated by C) |
| T | RELAPPG | I is mentioned in a relative clause or appositive referring to the phosphorylation (phosphorylation of A, which increased the interaction with B) |
The type column specifies if the feature is used in the detection of the temporal relation (T), causal relation (C) or both (T, C). The feature column lists the features by name and the description column gives a description of each feature. ‘P’ is short for phosphorylation and ‘I’ is short for interaction.
Features used in the detection of phosphorylation-interaction relations
| Type . | Feature . | Description . |
|---|---|---|
| T, C | SSI | Substrate is the same as interactant |
| T | IMP | One of the interactants is mentioned as being ‘phosphorylated’ (phosphorylated A binds to B) |
| T | CONJ | P and I are mentioned in a conjunction (there are five types of conjunctions captured in five different features) |
| C | ACTION | P and I are mentioned in a Subject–Verb–Object relationship (A phosphorylation leads to interaction with B) |
| T, C | DEPEND | I mentioned to be dependent on P (phosphorylation-dependent interaction of A to B) |
| T | PFIRST | P mentioned before I in the sentence |
| T | IFIRST | I mentioned before P in the sentence |
| T | WLR | There is a word/phrase between P and I hinting to a directionality of events from left to right (leads to) |
| T | WRL | There is a word/phrase between P and I hinting to a directionality of events from right to left (requires) |
| C | NEG | One of the events or the action is being negated (phosphorylated A does not bind to B) |
| C | HEDGE | One of the events or the action is mentioned with hedging (phosphorylated A might bind to B) |
| T | RELAPPB | P or I is mentioned in a relative clause or appositive referring to a protein (A, which interacts with B, is phosphorylated by C) |
| T | RELAPPG | I is mentioned in a relative clause or appositive referring to the phosphorylation (phosphorylation of A, which increased the interaction with B) |
| Type . | Feature . | Description . |
|---|---|---|
| T, C | SSI | Substrate is the same as interactant |
| T | IMP | One of the interactants is mentioned as being ‘phosphorylated’ (phosphorylated A binds to B) |
| T | CONJ | P and I are mentioned in a conjunction (there are five types of conjunctions captured in five different features) |
| C | ACTION | P and I are mentioned in a Subject–Verb–Object relationship (A phosphorylation leads to interaction with B) |
| T, C | DEPEND | I mentioned to be dependent on P (phosphorylation-dependent interaction of A to B) |
| T | PFIRST | P mentioned before I in the sentence |
| T | IFIRST | I mentioned before P in the sentence |
| T | WLR | There is a word/phrase between P and I hinting to a directionality of events from left to right (leads to) |
| T | WRL | There is a word/phrase between P and I hinting to a directionality of events from right to left (requires) |
| C | NEG | One of the events or the action is being negated (phosphorylated A does not bind to B) |
| C | HEDGE | One of the events or the action is mentioned with hedging (phosphorylated A might bind to B) |
| T | RELAPPB | P or I is mentioned in a relative clause or appositive referring to a protein (A, which interacts with B, is phosphorylated by C) |
| T | RELAPPG | I is mentioned in a relative clause or appositive referring to the phosphorylation (phosphorylation of A, which increased the interaction with B) |
The type column specifies if the feature is used in the detection of the temporal relation (T), causal relation (C) or both (T, C). The feature column lists the features by name and the description column gives a description of each feature. ‘P’ is short for phosphorylation and ‘I’ is short for interaction.
The binary features specific to this task capture both syntactic and lexical information about the phosphorylation and interaction. Some features are easily extracted (e.g. PFIRST, LFIRST and IMP), whereas others require more complicated analyses (e.g. SSI, CONJ and ACTION).
For example, in determining the SSI feature (substrate same as interactant), the interaction information alone is not sufficient. Contrast the following two sentences:
PAK phosphorylatesBad in vitro and in vivo on Ser112 and Ser136, resulting in a markedly reduced interaction between Bad and Bcl-2 or Bcl-x(L). (PMID 10611223)
Pim phosphorylation of Bad was also found to blockitsassociation with Bcl-XL. (PMID 16403219)
In the first sentence, both the substrate and one of the interactants are identified as ‘Bad’. The two occurrences being the same, it is straightforward to assign a value of 1 to the SSI feature. However, in the second sentence, the substrate is reported to be ‘Bad’, while one of the interactants is reported to be ‘its’. We look at the construction of the sentence to identify to which protein ‘its’ refers (in this case, it is ‘Bad’ that ‘its’ refers to, and the SSI feature is marked with a value of 1). Because the rules designed for the detection of PPIs are simple and do not cover all possible syntactic constructions, some complications can also arise in the detection of the SSI feature when one of the interactants is not explicitly defined, like in this example:
Serine phosphorylation of Bad is associated with 14-3-3binding. (PMID 11723239)
Thus, we need to identify the implicit interactant from previous protein mentions (i.e. substrate ‘Bad’ is the other interactant in this case).
There are five CONJ features (P and I are mentioned in a conjunction), marking five different types of coordination: (i) NP coordination involving the phosphorylation and interaction linked by ‘and’ (CONJ_NP), (ii) verb phrase coordination involving the two linked by ‘and’ (CONJ_VP), (iii) prepositional phrase coordination involving the phosphorylation and interaction linked by ‘and’ (CONJ_PP), (iv) sentential coordination involving the phosphorylation and interaction (CONJ_S) and (v) all other types of coordination (CONJ_O). These types of coordination are identified using a sentence simplifier developed in-house (27). Based on the type of coordination, a temporal relation can sometimes be determined between the phosphorylation and the interaction. Contrast the following two sentences containing coordination, the first hinting at a temporal relation, while the second does not.
Upon BCR activation, LAB is phosphorylated and interacts with Grb2. (PMID 15477350)
Contraction is associated with phosphorylation of myosin and interaction of actin with myosin. (PMID 20501443)
The phosphorylation or interaction appearing in a relative clause or appositive can also hint at no temporal relationship, and consequently no causal relationship, as can be seen in the example below (‘p-Bad’ in this case).
KD increased the protein protein interaction between 14-3-3 and p-Bad (Ser136), which might be phosphorylated by p-Akt (Ser473). (PMID 17058267)
However, if the relative clause or appositive refers to the phosphorylation event itself, then a temporal relationship can be determined. A sample sentence is shown here.
Akt1 mediated the phosphorylation of Bad at serine 136, which increased the interaction of serine 136-phosphorylated Bad with 14-3-3 proteins. (PMID 17555943)
Features such as PFIRST, IFIRST, WLR, WRL and the CONJ features are not sufficient on their own to point to a temporal relation. However, a combination of them can give us the temporal aspect (e.g. PFIRST + WLR, or IFIRST + WRL or PFIRST + CONJ_V).
Example rules are given below:
If SSI and PART OF, then mark with high confidence.
If SSI and DEPEND, then mark with high confidence.
If SSI, PFIRST, WRL, then mark with high confidence.
If SSI, ACTION, then mark with medium confidence.
If SSI, CONJ_V, PFIRST, then mark with medium confidence.
If SSI, PFIRST, CONJ, then mark with low confidence.
If SSI, IFIRST, WLR, then mark as negative.
Web interface and user interaction
To broaden the utility of eFIP for biocuration and knowledge discovery by biologists, a web interface is developed for users and biocurators to gather, modify and save literature mining results. eFIP’s results (obtained using the modules described above) are pre-processed and stored in a local database. Users can access the results online by searching for a gene/protein or by providing a list of PMIDs.
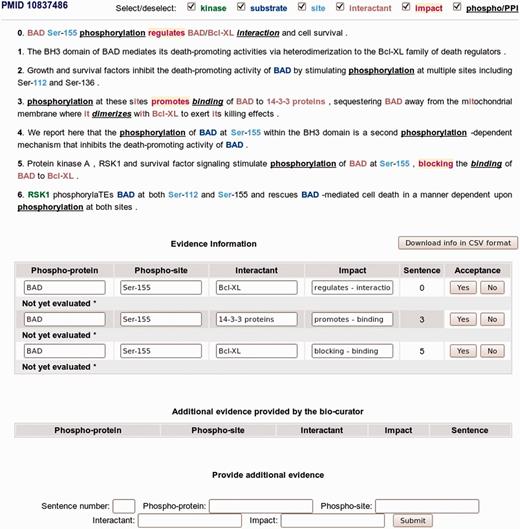
For any given protein, a ranked list of PMIDs is displayed, similar to the screenshot shown in Figure 2 for protein Bad. eFIP ranks these papers, taking into consideration the rules that applied to the sentences of an abstract and their confidence scores. If a list of PMIDs is provided as input, then multiple phosphorylated proteins might be involved in protein interactions. eFIP lists relevant PMIDs before the irrelevant ones, considering the number of phosphorylation and PPI mentions in the documents provided. To see the textual evidence in detail, a PMID can be selected from the ranked list and a page similar to the screenshot in Figure 3 is displayed for that PMID (in this case, PMID 10837486). The abstract is broken into sentences, and information for phosphorylation, interaction and impact is highlighted, where applicable.
eFIP ranking and result summary of abstracts for protein BAD. A total of 1331 abstracts are linked to protein BAD as determined by eGRAB, among which 369 mention phosphorylation information (ranked and partially shown). The ‘Impact’, ‘PPI’ and ‘Site’ images on the left point to the type of information are found in the abstract. The title, authors and a summary of the interactions involving the phosphorylated forms of BAD are displayed. A spreadsheet summary file can be downloaded by clicking on the ‘Download info in CSV format’ button.
eFIP annotation interface with sentence evidence attribution of phosphorylated protein and interaction events in PMID 10837486.
As eFIP aims to help biocurators and researchers find information about networks and functional impact of phosphorylated forms of proteins of their interest, we allow users of eFIP to log in, make corrections to the eFIP results, add evidence for sentences missed by eFIP and download the updated information. These corrections and additions of information will be saved only for that specific user, thus allowing multiple curators to work on the same abstracts at a time. The updated results can be downloaded for any given PMID by clicking ‘Download info in CSV format’ from the PMID page, and for any given ranked list of PMIDs by clicking the same button from the protein page. eFIP has been integrated into the PRO curation workflow and is used by PRO curators to capture validated text mining results and knowledge about phosphorylated forms of proteins.
Evaluation metrics
where true positive (TP) is the number of documents/sentences correctly found to be positive by eFIP, true negative (TN) is the number of documents/sentences correctly found by eFIP to be negative, false positive (FP) is the number of documents/sentences that eFIP mistakenly tags as positive and false negative (FN) is the number of documents/sentences that eFIP misses to tag as positive.
Results
The eFIP system participated in the BioCreative-2012 Workshop Track III—Interactive Text Mining. The requirements for this task included: (i) the system should be used in a biocuration workflow and have been evaluated internally and (ii) the system should have a web interface for users, with clear examples of input (such as PMID, gene and keyword) and output (list of relevant articles, compound recognition, PPI, etc.). Various evaluations have been conducted to measure the performance of eFIP for document retrieval, sentence-level information extraction, document ranking, as well as the utility and usability of the eFIP web interface for biocurators. We will first describe the data sets used in the evaluations, and then provide results for each evaluation.
Data sets
Two types of data sets were constructed: one for a user evaluation of eFIP during BioCreative-2012 Workshop, and the other for an in-house evaluation of the system.
The BioCreative-2012 user evaluation involved two curators recruited by the BioCreative organizers. The curators are domain experts from Merck and Reactome. To conduct a document-centric evaluation, 50 abstracts were randomly selected based on proteins involved in two pathways of interest to Reactome (i.e. autophagy and HIV infection). A boolean query in PubMed was constructed using the keywords ‘autophagy’ or ‘HIV’ in combination with phosphorylat* and several derivatives of the interaction verbs. The two curators were assigned the same 25 abstracts to curate manually (BioCreative Set 1) and 25 abstracts to curate using the eFIP interface (BioCreative Set 2). For a protein-centric evaluation, BioCreative-2012 provided four additional sets of abstracts consisting of top 10 abstracts for the following proteins: PLCG1, LAT, LCP2 and ZAP70 (BioCreative Set 3). These proteins were chosen randomly from the proteins involved in the adaptive immune system (Reactome: REACT_75774). We note here that these evaluations have been shown to be time consuming, thus the number of abstracts chosen to be included in the evaluation was determined based on the curators’ availability.
Because the BioCreative-2012 data sets were constructed around certain proteins and pathways, and thus do not necessarily capture the broad spectrum of abstracts that are relevant for eFIP, we therefore conducted a system evaluation to benchmark how well eFIP performs on a broad set of proteins and abstracts mentioning phosphorylation and interaction events (In-house Set). We randomly selected 96 abstracts (distinct from the set used in the development of eFIP) from all PubMed abstracts containing sentences with trigger words for both phosphorylation and interaction mentions. The test set was manually curated by PRO curators without the use of the eFIP interface.
The set of abstracts represents a gold standard literature corpus and is made publically available through the iProLINK website at http://proteininformationresource.org/pirwww/iprolink/eFIPCorpus.txt. For each abstract, we list the PMID, the original title and abstract, as well as the individual sentences coming from the abstract. Where applicable, these sentences are further marked with phosphorylated protein, kinase, site, interactant, type of interaction and effect.
Annotation process
For the manual curation (i.e. BioCreative Set 1 and In-house Set), we asked the biocurators to read the abstracts in PubMed and gather, in a spreadsheet, both phosphorylation information (phosphorylated protein and site) and interaction information (the interactants, the specific word(s) pointing to the interaction and the impact on the interaction), only from the sentences in which there was a clear indication that the phosphorylated protein (substrate) is involved in the interaction. Note that for this manual curation, the curators were not allowed to interact with eFIP or see the system’s results.
For the evaluation involving the use of the eFIP annotation interface (i.e. BioCreative Set 2 and BioCreative Set 3), the biocurators were asked to log in and mark as relevant or irrelevant the results proposed by eFIP, and to add any other textual evidence that was missed by eFIP. This provided user evaluation on how useful the eFIP interface is, and on time it takes to find information using eFIP versus reading abstracts in PubMed.
Recall that BioCreative Set 1 and BioCreative Set 2 were annotated by two curators independently. We examined the inter-annotator agreement and asked for the opinion of a PRO curator for the cases in which they disagreed. Except for a few instances (7 disagreements), the two curators agreed on their common annotations (54 agreements). In a few cases, however, we noticed that the annotators did not completely follow the guidelines of annotating every relevant sentence whether it contained redundant information. There were eight such ‘redundant’ sentences in the entire set. While one annotator extracted information in all relevant sentences, the other annotator marked the information from only one of the relevant sentences and not those with the redundant information. Thus, we present two inter-annotator agreement scores: (i) sentence level: based on the sentences annotated by both annotators and (ii) fact level: based on the annotated facts that were extracted from these abstracts. The Kappa coefficient for inter-annotator agreement was 0.80 at the sentence level and 0.77 at the fact level. This is considered as a significant agreement [a Kappa coefficient of 0.61 or above is considered to be a substantial agreement in the literature (28)].
The annotation guidelines for eFIP evaluation can be accessed from the iProLINK website at http://proteininformationresource.org/pirwww/iprolink/eFIP-annotation-guidelines.pdf.
Evaluating eFIP on document retrieval
A main goal of eFIP is to suggest documents containing information that is relevant for biocuration. For this, we conducted system evaluation of eFIP in terms of precision (P), recall (R) and F-measure (F) at the document level by running the eFIP in batch mode against the four manually curated data sets. The results are shown in Table 3 for the In-house Set of randomly picked abstracts (71.1% P, 86.5% R and 78% F), BioCreative Set 1 of manually curated abstracts (100% P/R/F), BioCreative Set 2 of abstracts curated using the eFIP interface (83.3% P/R/F) and BioCreative Set 3 of top 10 abstracts for four different proteins (82.4% P, 93.3% R and 87.5% F).
eFIP performance evaluation on document retrieval as measured by precision, recall and F-measure based on TP, TN, FP and FN
| Evaluation set . | # Abstracts . | Precision . | Recall . | F-measure . | TP . | TN . | FP . | FN . |
|---|---|---|---|---|---|---|---|---|
| In-house Set | 96 | 71.1 | 86.5 | 78.0 | 32 | 46 | 13 | 5 |
| BioCreative Set 1 | 25 | 100.0 | 100.0 | 100.0 | 11 | 14 | 0 | 0 |
| BioCreative Set 2 | 25 | 83.3 | 83.3 | 83.3 | 10 | 11 | 2 | 2 |
| BioCreative Set 3 | 40 | 82.4 | 93.3 | 87.5 | 28 | 4 | 6 | 2 |
| Evaluation set . | # Abstracts . | Precision . | Recall . | F-measure . | TP . | TN . | FP . | FN . |
|---|---|---|---|---|---|---|---|---|
| In-house Set | 96 | 71.1 | 86.5 | 78.0 | 32 | 46 | 13 | 5 |
| BioCreative Set 1 | 25 | 100.0 | 100.0 | 100.0 | 11 | 14 | 0 | 0 |
| BioCreative Set 2 | 25 | 83.3 | 83.3 | 83.3 | 10 | 11 | 2 | 2 |
| BioCreative Set 3 | 40 | 82.4 | 93.3 | 87.5 | 28 | 4 | 6 | 2 |
eFIP performance evaluation on document retrieval as measured by precision, recall and F-measure based on TP, TN, FP and FN
| Evaluation set . | # Abstracts . | Precision . | Recall . | F-measure . | TP . | TN . | FP . | FN . |
|---|---|---|---|---|---|---|---|---|
| In-house Set | 96 | 71.1 | 86.5 | 78.0 | 32 | 46 | 13 | 5 |
| BioCreative Set 1 | 25 | 100.0 | 100.0 | 100.0 | 11 | 14 | 0 | 0 |
| BioCreative Set 2 | 25 | 83.3 | 83.3 | 83.3 | 10 | 11 | 2 | 2 |
| BioCreative Set 3 | 40 | 82.4 | 93.3 | 87.5 | 28 | 4 | 6 | 2 |
| Evaluation set . | # Abstracts . | Precision . | Recall . | F-measure . | TP . | TN . | FP . | FN . |
|---|---|---|---|---|---|---|---|---|
| In-house Set | 96 | 71.1 | 86.5 | 78.0 | 32 | 46 | 13 | 5 |
| BioCreative Set 1 | 25 | 100.0 | 100.0 | 100.0 | 11 | 14 | 0 | 0 |
| BioCreative Set 2 | 25 | 83.3 | 83.3 | 83.3 | 10 | 11 | 2 | 2 |
| BioCreative Set 3 | 40 | 82.4 | 93.3 | 87.5 | 28 | 4 | 6 | 2 |
Evaluating eFIP on sentence-level information extraction
Because eFIP also suggests sentences and exact information from these sentences for biocuration, we also conducted an evaluation of the information extracted at a sentence level. The results are calculated in terms of precision, recall and F-measure, and are shown in Table 4 for the system evaluation (eFIP batch processing) of In-house Set (72.4% P, 67.9% R and 70.1% F), and the user evaluation (interactive mode using eFIP interface) of BioCreative Set 2 (94.7% P, 69.2% R and 80% F), in contrast to the manual curation (without using eFIP) of BioCreative Set 1 (84.2% P, 80% R and 82% F).
eFIP performance evaluation on information extraction at the sentence level as measured by precision, recall and F-measure based on TP, TN, FP and FN
| Evaluation type . | # Abstracts/sentences . | Time to completion . | Precision . | Recall . | F-measure . | TP . | TN . | FP . | FN . |
|---|---|---|---|---|---|---|---|---|---|
| System evaluation (In-house) | 96/148 | 72.4 | 67.9 | 70.1 | 55 | 46 | 21 | 26 | |
| User evaluation (BioCreative) | |||||||||
| Set 1: Manual curation | 25/37 | 104 min | 84.2 | 80.0 | 82.0 | 16 | 14 | 3 | 4 |
| Set 2: eFIP interface | 25/37 | 42.5 min | 94.7 | 69.2 | 80.0 | 18 | 10 | 1 | 8 |
| Evaluation type . | # Abstracts/sentences . | Time to completion . | Precision . | Recall . | F-measure . | TP . | TN . | FP . | FN . |
|---|---|---|---|---|---|---|---|---|---|
| System evaluation (In-house) | 96/148 | 72.4 | 67.9 | 70.1 | 55 | 46 | 21 | 26 | |
| User evaluation (BioCreative) | |||||||||
| Set 1: Manual curation | 25/37 | 104 min | 84.2 | 80.0 | 82.0 | 16 | 14 | 3 | 4 |
| Set 2: eFIP interface | 25/37 | 42.5 min | 94.7 | 69.2 | 80.0 | 18 | 10 | 1 | 8 |
eFIP performance evaluation on information extraction at the sentence level as measured by precision, recall and F-measure based on TP, TN, FP and FN
| Evaluation type . | # Abstracts/sentences . | Time to completion . | Precision . | Recall . | F-measure . | TP . | TN . | FP . | FN . |
|---|---|---|---|---|---|---|---|---|---|
| System evaluation (In-house) | 96/148 | 72.4 | 67.9 | 70.1 | 55 | 46 | 21 | 26 | |
| User evaluation (BioCreative) | |||||||||
| Set 1: Manual curation | 25/37 | 104 min | 84.2 | 80.0 | 82.0 | 16 | 14 | 3 | 4 |
| Set 2: eFIP interface | 25/37 | 42.5 min | 94.7 | 69.2 | 80.0 | 18 | 10 | 1 | 8 |
| Evaluation type . | # Abstracts/sentences . | Time to completion . | Precision . | Recall . | F-measure . | TP . | TN . | FP . | FN . |
|---|---|---|---|---|---|---|---|---|---|
| System evaluation (In-house) | 96/148 | 72.4 | 67.9 | 70.1 | 55 | 46 | 21 | 26 | |
| User evaluation (BioCreative) | |||||||||
| Set 1: Manual curation | 25/37 | 104 min | 84.2 | 80.0 | 82.0 | 16 | 14 | 3 | 4 |
| Set 2: eFIP interface | 25/37 | 42.5 min | 94.7 | 69.2 | 80.0 | 18 | 10 | 1 | 8 |
Overall, the evaluation using the eFIP interface achieved better precision (94.7 versus 84.2%), lower recall (69.2 versus 80%) and similar F-measure (80 versus 82%). The most significant outcome of using the eFIP interface is time to completion for the curation task. For both curators, annotation using the eFIP system took significantly less time than the manual curation (from 120 to 50 min for the first curator and from 88 to 35 min for the second curator). This averages to 104 min for manual curation and 42.5 min for eFIP-based curation, an ∼2.5-fold speed-up with the usage of the eFIP text mining interface.
Evaluating eFIP on document ranking
The eFIP system ranks abstracts based on the amount of relevant information they contain. As both document prioritization and information extraction are important factors in speeding up the curation process, we therefore evaluated how well eFIP can prioritize documents for curation. The results are shown in Table 5 in terms of normalized discounted cumulative gain (nDCG) for the In-house Set (94.5%), BioCreative Set 1 (100%), BioCreative Set 2 (98.08%) and BioCreative Set 3, i.e., the top 10 abstracts for each individual protein chosen during the BioCreative-2012 evaluation (LAT 100%, LCP2 98.76%, PLCG1 93.45% and ZAP70 96.2%). The average precision (AveP) is also shown in Table 5 for these sets, giving a MAP of 0.86.
eFIP performance evaluation on document ranking as measured by nDCG and AveP based on the ranked lists of abstracts
| Evaluation set . | # Abstracts . | Relevant . | Irrelevant . | nDCG . | AveP . |
|---|---|---|---|---|---|
| In-house Set | 96 | 37 | 59 | 94.50 | 0.75 |
| BioCreative Set 1 | 25 | 11 | 14 | 100.00 | 1.00 |
| BioCreative Set 2 | 25 | 12 | 13 | 98.08 | 0.81 |
| Protein | |||||
| LAT | 10 | 10 | 0 | 100.00 | 1.00 |
| LCP2 | 10 | 8 | 2 | 98.76 | 0.83 |
| PLCG1 | 10 | 4 | 6 | 93.45 | 0.73 |
| ZAP70 | 10 | 8 | 2 | 96.20 | 0.88 |
| Evaluation set . | # Abstracts . | Relevant . | Irrelevant . | nDCG . | AveP . |
|---|---|---|---|---|---|
| In-house Set | 96 | 37 | 59 | 94.50 | 0.75 |
| BioCreative Set 1 | 25 | 11 | 14 | 100.00 | 1.00 |
| BioCreative Set 2 | 25 | 12 | 13 | 98.08 | 0.81 |
| Protein | |||||
| LAT | 10 | 10 | 0 | 100.00 | 1.00 |
| LCP2 | 10 | 8 | 2 | 98.76 | 0.83 |
| PLCG1 | 10 | 4 | 6 | 93.45 | 0.73 |
| ZAP70 | 10 | 8 | 2 | 96.20 | 0.88 |
eFIP performance evaluation on document ranking as measured by nDCG and AveP based on the ranked lists of abstracts
| Evaluation set . | # Abstracts . | Relevant . | Irrelevant . | nDCG . | AveP . |
|---|---|---|---|---|---|
| In-house Set | 96 | 37 | 59 | 94.50 | 0.75 |
| BioCreative Set 1 | 25 | 11 | 14 | 100.00 | 1.00 |
| BioCreative Set 2 | 25 | 12 | 13 | 98.08 | 0.81 |
| Protein | |||||
| LAT | 10 | 10 | 0 | 100.00 | 1.00 |
| LCP2 | 10 | 8 | 2 | 98.76 | 0.83 |
| PLCG1 | 10 | 4 | 6 | 93.45 | 0.73 |
| ZAP70 | 10 | 8 | 2 | 96.20 | 0.88 |
| Evaluation set . | # Abstracts . | Relevant . | Irrelevant . | nDCG . | AveP . |
|---|---|---|---|---|---|
| In-house Set | 96 | 37 | 59 | 94.50 | 0.75 |
| BioCreative Set 1 | 25 | 11 | 14 | 100.00 | 1.00 |
| BioCreative Set 2 | 25 | 12 | 13 | 98.08 | 0.81 |
| Protein | |||||
| LAT | 10 | 10 | 0 | 100.00 | 1.00 |
| LCP2 | 10 | 8 | 2 | 98.76 | 0.83 |
| PLCG1 | 10 | 4 | 6 | 93.45 | 0.73 |
| ZAP70 | 10 | 8 | 2 | 96.20 | 0.88 |
Note that the eFIP system significantly simplifies the task of finding relevant articles in the literature. This is reflected in the number of relevant documents found by eFIP compared with the total number of documents mentioning the given protein (determined by eGRAB), or compared with the number of articles containing phosphorylation mention (determined by RLIMS-P). For example, 507 abstracts mention protein LAT, but only 125 of these contain mentions of phosphorylation and only 19 are marked by eFIP as containing phosphorylation–PPI relations. Similar distributions are observed for the other three proteins (LCP2: 309-96-9, PLCG1: 676-100-5 and ZAP70: 1105-181-25).
Evaluating eFIP on usability
eFIP was also evaluated by biocurators during the BioCreative-2012 Workshop demo session. Each evaluator provided answers to a questionnaire regarding their experience using the eFIP system. The overall feedback was highly positive. On a scale from 1 to 7 from lowest to highest, eFIP scored at 6. Users liked the ability to correct and download results on the fly, the color-coded highlighting of different entities and the display of information content for ranked abstracts. Improvements of the system and the online interface were suggested during the demo session, some of which we have already taken into consideration.
Discussion
The eFIP evaluation has shown that the system performs well at its task, in particular in regard to document retrieval, ranking and usability. In this section, we discuss some issues based on an analysis of its errors in the sentence-level information extraction evaluation.
We have developed eFIP as an end-to-end system. Any error in part-of-speech tagging, parsing or by one of the components (e.g. RLIMS-P or the PPI module) will likely cause an eFIP error. In fact, around 58% of the eFIP errors (FN or FP) can be directly attributed to RLIMS-P or the PPI module's errors, and most of the remaining errors can be attributed to the impact module. When we manually correct these errors in the In-house Set abstracts, the eFIP's precision, recall and F-measure go up to 75.9, 81.5 and 78.6% from 72.4, 67.9 and 70.1%, respectively. Development of tools for extracting post-translational modification and PPI information (13, 29) are active research topics, and any improvement we make to these two tools will directly translate into furthering the accuracy of eFIP.
A third of the FPs (contributing to a drop in precision) is due to mistakes of the PPI module. We attribute of the remaining FPs to complications in the detection of temporal relations or due to the selection of sentences that describe experimental setups rather than actual results:
Both CAD and phosphorylatedKID have been proposed to recruitpolymerase complexes, but this has not been directly tested. (PMID 11158288)
One reason for FNs (contributing to a drop in recall) is due to the PPI module being unable to identify an interaction event. For example, consider the following sentence in which the trigger word ‘binding’ occurs, but no interacting proteins could be detected for the binding:
In addition, we provide evidence that phosphorylation of the splice variant region is unlikely to represent the mechanism by which binding is reduced. (PMID 15225631)
Because the PPI module requires at least one interactant to be identified for the trigger word (e.g. ‘binding’), this sentence is automatically marked as negative. A similar situation happens when RLIMS-P fails to detect phosphorylation events.
Failure to detect that one of the interactants is the same as the phosphorylated protein (see the SSI feature in Table 2) was a cause for a few FNs. This can happen when the PPI module identifies the wrong interactant, or when the sentence is written in a way that is ambiguous for the detection.
FNs are also partly due to the complexity of some sentences. Sentences with complex grammatical structures are observed particularly in the abstracts of scientific articles, as authors try to summarize, in a few sentences, the various facts described throughout the manuscript. For example, consider the following sentence:
Here we discovered that phosphorylation of Ser(88), which juxtapose each other at the interface of the DLC, disrupts DLC1 dimer formation and consequently impairs its interaction with Bim. (PMID 18084006)
This sentence contains a relative clause, emphasized in italics, which stands in the way of detecting that the phosphorylation disrupts the dimer formation. If we can somehow skip over the relative clause, then the syntax becomes simple [‘Phosphorylation of Ser(88) disrupts DLC1 dimer formation’] and will match one of the PPI patterns. Additionally, this sentence contains a coordination involving the two PPI mentions (i.e. ‘dimer formation’ and ‘interaction’). If we can detect that the two mentions are part of a coordination and skip over the first conjunct (i.e. ‘disrupts DLC1 dimer formation’), then we would be left with a simple sentence matching a PPI pattern [‘Phosphorylation of Ser(88) consequently impairs its interaction with Bim’].
Several sentence simplifiers have been suggested for the biomedical text, and we plan to incorporate one such simplifier in the eFIP pipeline. The use of a sentence simplifier has been reported to drastically improve the performance of biomedical text mining and relation extraction systems (27,30).
Conclusion and future work
In this article, we have described a system, eFIP, for detecting literature relevant to PPIs involving phosphorylated proteins. eFIP integrates eGRAB and RLIMS-P for the retrieval of all documents relevant to a particular phosphorylated protein, and a PPI module that relies on syntactic patterns to extract interacting partners. The impact module uses a rule-based approach that detects relations between phosphorylation and interaction events, as well as impact of phosphorylation on interaction where applicable.
Several new functionalities are being added to eFIP to facilitate knowledge discovery from phosphorylation protein networks that connect phosphorylated proteins with kinases and interaction partners, along with their functional outcomes. We will integrate kinase information extracted from RLIMS-P to the eFIP text mining summary tables and results for kinase-substrate relationships. Furthermore, because phosphorylation of a protein can have an impact on not only its interaction with other proteins but also the regulation of its molecular function (such as activity) and subcellular localization, we will further explore the detection of these types of impact. We plan to incorporate eGIFT (3) in the pipeline of eFIP for the detection of molecular functions, biological processes and subcellular localizations relevant to a phosphorylated protein. Third, the PPI module currently handles a limited set of interaction types (i.e. affinity, association, binding, complex, disassociation, dimerization, interaction and recruitment). In the future, we plan to extend the PPI module to include additional types of interactions, such as co-precipitation, release and sequestering.
We will improve the performance of the eFIP text mining with several enhancements. eFIP’s rules are currently focused on single sentences. In the future, we will extend the rules to detect phosphorylation-interaction relations that span multiple sentences. Although the Results section shows that the manually designed rules detect the existence of relevant information with high accuracy, we still want to explore how machine learning could improve the performance of eFIP when the same set of features is used on a larger set of annotated abstracts. We also plan to use a new sentence simplifier, iSimp (31), to improve the recall of the impact module of eFIP. The incorporation of sentence simplifiers in the pipeline of eFIP is expected to result in more comprehensive detection of the effect of phosphorylation on the interaction.
Funding
This work was supported by National Science Foundation grant ABI-1062520 and National Institutes of Health grants 5G08LM010720-02 and 5R01GM080646-06. Funding for open access charge: National Science Foundation grant ABI-1062520.
Conflict of interest. None declared.
Acknowledgements
We would like to thank the BioCreative organizing committee for providing the venue for eFIP user evaluation, and the curators who took time to participate in the testing and evaluation of eFIP.