-
PDF
- Split View
-
Views
-
Cite
Cite
Alexander Junge, Jan C. Refsgaard, Christian Garde, Xiaoyong Pan, Alberto Santos, Ferhat Alkan, Christian Anthon, Christian von Mering, Christopher T. Workman, Lars Juhl Jensen, Jan Gorodkin, RAIN: RNA–protein Association and Interaction Networks, Database, Volume 2017, 2017, baw167, https://doi.org/10.1093/database/baw167
Close - Share Icon Share
Abstract
Protein association networks can be inferred from a range of resources including experimental data, literature mining and computational predictions. These types of evidence are emerging for non-coding RNAs (ncRNAs) as well. However, integration of ncRNAs into protein association networks is challenging due to data heterogeneity. Here, we present a database of ncRNA–RNA and ncRNA–protein interactions and its integration with the STRING database of protein–protein interactions. These ncRNA associations cover four organisms and have been established from curated examples, experimental data, interaction predictions and automatic literature mining. RAIN uses an integrative scoring scheme to assign a confidence score to each interaction. We demonstrate that RAIN outperforms the underlying microRNA-target predictions in inferring ncRNA interactions. RAIN can be operated through an easily accessible web interface and all interaction data can be downloaded.
Database URL:http://rth.dk/resources/rain
Introduction
The study of protein-coding genes and the accumulation of data from expression studies and other complementary methods have helped researchers to generate protein association networks compiled in resources such as the STRING database (1). Using a probabilistic scoring scheme, STRING assigns a score to each physical interaction and functional association (henceforth referred to as interactions). The recent version 10 holds interactions for >2000 organisms.
However, interaction networks containing only proteins and their interactions remain incomplete until other important molecular interactions have been included. For this reason, we have focused on complementing protein interaction networks with non-coding RNAs (ncRNAs)—a large class of genes comprising ∼16 000 long and ∼10 000 short ncRNAs in human [GENCODE version 24 (2)]. Integration of these interactions allows for an analysis of the complex functional interplay of ncRNA–RNA and ncRNA–protein interactions. Data on such interactions, complemented by co-expression and literature mining, are currently emerging (3–5). This led to the generation of databases storing ncRNA interactions such as miRTarBase (6) and TarBase (7) containing microRNA (miRNA)–target interactions. NPInter (5), RAID (8) and StarBase (9) are examples of databases collecting interactions between ncRNAs and proteins.
The analysis of ncRNA interactions is challenged by issues related to data heterogeneity, such as varying quality as well as the usage of different identifiers and interaction scoring schemes. The STRING database, used by thousands of researchers daily, has addressed these challenges for proteins through the use of unified identifiers and calibrated scoring schemes (1). A resource similar to STRING is not available for ncRNAs and their interactions.
Similar to protein interactions, ncRNA interactions are supported by diverse sources of evidence such as expert curation, experiments, text mining and predictions. In order to compare these sources of evidence, a scoring scheme needs to be established that assesses the reliability of each interaction. NcRNAs interacting with either proteins or ncRNAs furthermore affect the pathways these interaction partners are involved in. Hence, an approach that makes it easy to navigate both ncRNA as well as protein association networks promises to benefit the study of cellular interaction networks.
We have used a strategy similar to that of STRING in order to develop RAIN (RNA–protein Association and Interaction Networks), a novel resource that covers ncRNA and their associations with other ncRNAs and proteins. RAIN integrates ncRNA interactions from a diverse set of sources and covers four organisms: human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus) and baker’s yeast (Saccharomyces cerevisiae). RAIN scores the reliability of each interaction using a scoring scheme based on the comparison to a curated set of interactions. It finally integrates ncRNA–RNA and ncRNA–protein associations with protein–protein associations contained in the STRING database. This enables researchers to explore complex interaction networks in the powerful, yet intuitive interactive STRING user interface.
Materials and Methods
Sources of evidence
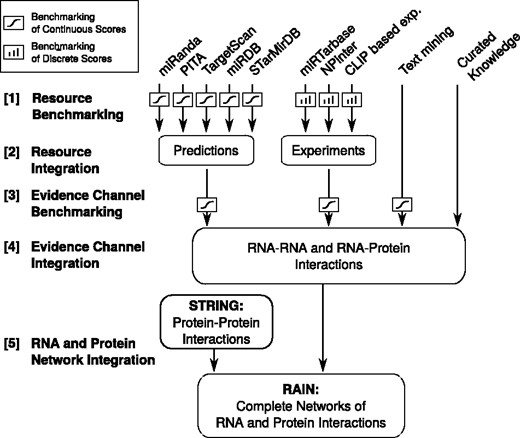
We established four channels of evidence to support the interactions found in RAIN, namely, (i) curated knowledge, (ii) experimental evidence, (iii) miRNA target predictions and (iv) automated literature mining, see Figure 1. Each of the four evidence channels is generated by integrating a number of underlying resources.
Flow chart illustrating the development of the RAIN database, ranging from establishing scoring schemes for the individual sources of evidence, through integration of resources to evidence channels, to finally defining functional molecular networks.
(i) Curated knowledge. This comprises 867 human molecular interactions that are well established in the scientific literature and/or listed in expert curated databases. The interactions were collected for nine classes of ncRNAs, namely microRNA (miRNA) (3), ribosomal RNA (rRNA) (10), transfer RNA (tRNA) (11), signal recognition particle RNA (SRP RNA) (12), Vault RNA (13–15), Y RNA (16–18), Telomerase RNA (19), small nucleolar RNA (snoRNA) (20) and spliceosomal RNA (U1, U2, U4, U4atac, U6, U6atac, U11, U12) (20). For further details on the curated interactions, refer to Supplementary Section 2.
(ii) Experimental evidence. This comprises 10 588 interactions supported by experimental data. Cross-linking immunoprecipitation (CLIP) based experiments were retrieved from StarBase (9) and supplemented by interactions identified in CLASH and CRAC experiments (21–23). Furthermore, experimentally supported interactions were extracted from miRTarBase (6) and NPInter (5) and redundancy between the databases was removed. The confidence of the experimental evidence was based on the number of experiments supporting a given interaction.
(iii) miRNA target predictions: We ran miRanda (24) and PITA (25) with default settings on all combinations of 3′ UTR sequences of protein-coding genes from Ensembl Biomart (26) and miRNA sequences from miRBase (27). Additionally, we retrieved precomputed predictions for miRDB (28), TargetScan (29, 30) and StarMirDB (31).
(iv) Text mining. ncRNA orthology groups were generated using Ensembl Biomart (26) and the miRNA family annotations from miRBase v20 (27). Protein orthology groups retrieved from STRING and these ncRNA orthology groups were supplied to the dictionary-based named entity recognition engine described by Pafilis et al. (32) to extract associations between ncRNAs and proteins from MEDLINE abstracts. We refer to Pafilis et al. (32) for more details on the named entity recognition software. The subsequent text mining was performed using the same name tagger as used in STRING (33).
A confidence score is assigned to each evidence for an association. Curated associations were considered highly reliable and assigned the highest possible confidence score for a single source of evidence, defined as 0.9 in STRING. Experimentally supported associations were assigned confidence scores based on the number of supporting experiments/publications. As in STRING (33), associations derived from text mining were scored based on co-occurrences of gene names. For miRNA target predictions, we used the scoring schemes of the individual predictors, at the outset. To put these heterogeneous scores on a common scale, we converted them to probabilistic scores through benchmarking against the same gold standard set (Figure 1, Step [1]). Assuming independence between the sources of evidence, the combined probability of an association was computed from the resource-specific probabilistic scores (Figure 1, Step [2]). The combined probabilities were subjected to a second round of benchmarking to mitigate violations of the assumption of independence (Figure 1, Step [3]). Finally, the evidence channels were integrated to establish the ncRNA association networks (Figure 1, Step [4]) that interface with STRING to provide a complete ncRNA and protein interaction network (Figure 1, Step [5]). We restricted RAIN to only cover organisms with at least 500 ncRNA interactions with confidence scores > 0.15 (the same cutoff is used in STRING) which resulted in the inclusion of human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus) and baker’s yeast (Saccharomyces cerevisiae).
The gold standard set contained 782 miRNA–mRNA interactions that were deemed to be highly reliable. The interactions involve 171 miRNAs and 437 mRNAs. We defined our gold standard based on the curated miRNA–mRNA interactions from Croft et al. (3) as well as miRNA–mRNA interactions from miRTarBase (6) and NPInter (5) that were supported by at least two low-throughput experiments. We defined a low-throughput experiment as one that reports less than five miRNA interactions. To ensure an independent benchmarking of miRTarBase and NPInter, we excluded gold standard interactions originating from miRTarBase and NPInter while establishing the resource-specific probabilistic scoring scheme. Once fitted, this scoring scheme was applied to all interactions, including those defined as gold standard interactions.
Naming convention
A consistent naming convention in RAIN was achieved by compiling name and identifier aliases of ncRNA and proteins and generating an alias dictionary that maps these aliases to RAIN identifiers. For proteins and mRNA, RAIN identifiers are equivalent with STRING v10 (1) identifiers, and the alias dictionary is derived from the STRING v10 alias files. Aliases of miRNA were generated from miRBase v20 (27) and the associated miRBase identifiers were used subsequently. Finally, aliases of the remaining ncRNAs were retrieved using Ensembl Biomart v78 (26) and the official name of the given ncRNA was used as the RAIN identifier. The organism-specific database dictated these official names, i.e. HGNC (34) for human, MGI (35) for mouse and rat, and SGD (36) for yeast. All molecular entities were made to conform to the RAIN naming convention prior to establishing the probabilistic scoring schemes.
Probabilistic scoring schemes
For each resource of ncRNA–target interactions integrated into RAIN, a probabilistic scoring scheme was established prior to the process of resource integration. This allowed us to weight the respective resources based on their confidence in the final score integration step, which assigns an easily interpretable confidence score to each interaction.
The probabilistic scoring scheme is established by benchmarking against a gold standard set, Ω, of 782 miRNA–mRNA interactions that are considered to be valid. We denote the ith miRNA–mRNA interaction pair as ωi and thus . Let denote the set of all possible interactions between the miRNAs, , and the mRNAs, , contributing to the interactions in Ω. Hence, . An interaction between miRNA and mRNA contributed by an interaction resource is a true positive (TP) if . Similarly, is a false positive (FP) if .
This is summarized in Figure 2A: is the benchmark data set consisting of positive examples and negative examples . In Figure 2A, a white dot represents a TP interaction and a black dot represents a FP interaction. The universe of miRNA–mRNA interactions can be extended beyond covering miRNAs and miRNAs not present in and , respectively. We call this set and note that . Interactions in are represented by gray dots in Figure 2A.
Toy example describing the benchmarking and scoring scheme. (A) A true positive (TP) interaction is depicted as a black dot and represents a miRNA–mRNA pair found in the gold standard; a false positive (FP) interaction is depicted as a white dot and comprises interactions where the miRNA and mRNA constituents are in the gold standard, but their pair is not. Interactions where the miRNA or the mRNA were not part of the gold standard are depicted as gray dots. Only TP and FP interactions are used to establish the transfer function, which subsequently is applied to assign confidence scores to all interactions. (B) A discrete transfer function is established as the fraction of correctly predicted interactions in each of the discrete raw score bins. (C) A continuous transfer function is established based on the TP and FP interactions found in sliding windows. The mean raw interaction score and fraction of correctly predicted interactions were computed for each window, followed by the fitting of a sigmoid transfer function.
The following was performed in the interest of estimating whether a gray dot represents a likely interaction or not. Each ncRNA interaction resource had a discrete or continuous raw score assigned to each potential interaction contributed by the resource. To ensure that these scores were comparable, we calibrated them based on their agreement with the benchmark set . This calibration procedure is described in the following sections.
Scoring schemes for discrete raw scores
When a source of ncRNA interactions provides discrete raw scores, we calibrated by fitting a discrete transfer function as exemplified in Figure 2B. An example of such discrete scores are the interactions extracted from the portion of miRTarBase not overlapping our benchmark set . Here, the raw score was defined by the number of publications supporting a given interaction.
For each discrete raw score, the fraction of correctly predicted interactions, , was computed for the set of interactions with the given score. This provided a mapping of raw scores to the interval and defined the transfer function from the raw score assigned by a specific interaction resource to its confidence score , where estimates the probability that interactions assigned with the raw score are true.
Scoring schemes for continuous raw scores
After the fitting process, we applied f to map continuous raw scores to confidence scores , representing the probability of the interaction being true, as depicted in Figure 2C.
Integration of evidence
The prior probability is defined as the probability of randomly selecting a true positive miRNA–mRNA interaction from all combinations of and . Given our benchmarking set, the prior used in RAIN is . When computing , accounting for the prior is required to avoid counting the prior for each evidence channel. This prior correction is especially important when dealing with low score close to the prior (see Supplementary Section 5).
Following this integration of the sources of evidence into evidence channels, a second round of calibration was employed to mitigate any violations to the assumption of independence between interaction resources. Note that although each confidence score was computed based on a gold standard only consisting of miRNA–mRNA interactions, the underlying transfer functions mapping raw scores to confidence scores can be applied to score interactions for any class of ncRNAs.
Validation of the integration of miRNA target predictors
For the purpose of evaluating the gain of integrating the respective miRNA target prediction tools into the RAIN prediction channel, we retrieved a list of human and mouse miRNA–mRNA interactions from TarBase (7), that have been tested with functional studies (Luciferase reporter assays). All interactions common between this TarBase set and our gold standard were removed from the TarBase set to establish an independent validation set. This independent validation set comprises a positive set of 1387 confirmed interactions and a negative set of 460 pairs for which the miRNA had no effect on the amount of translated mRNA. The performance was assessed using receiver-operating characteristics (ROC) on the raw scores from the miRNA prediction tools and the combined probabilistic scores for the RAIN prediction channel.
Results and Discussion
RAIN is a novel resource of ncRNA interactions that integrates heterogeneous evidence from experiments, predictions, text mining and expert curation. RAIN comprises a total of 270 242 ncRNA–RNA/protein interactions across four widely investigated organisms: human, mouse, rat and yeast. The number of interactions is summarized in Table 1, with an additional break down of the counts by evidence channel and class of interacting entities in the Supplementary Tables S1 and S2. Furthermore, RAIN interfaces tightly with STRING (1) enabling users to explore networks of ncRNA–RNA, ncRNA–protein and protein–protein associations in an interactive user interface, with the reliability of each interactions represented as a single easily interpretable confidence score. RAIN is to our knowledge the first resource to offer this.
The number of miRNA–mRNA, ncRNA–protein and ncRNA–ncRNA interactions per organism in RAIN with a combined confidence score higher than 0.15
| Organism . | Number of interactions . | |||
|---|---|---|---|---|
| . | miRNA–mRNA . | ncRNA–protein . | ncRNA–ncRNA . | Total . |
| H. sapiens (human) | 174 853 | 11 026 | 2507 | 188 386 |
| M. musculus (mouse) | 77 270 | 469 | 35 | 77 774 |
| R. norvegicus (rat) | 19 985 | 39 | 1 | 20 025 |
| S. cerevisiae (Baker’s yeast) | 0 | 640 | 85 | 725 |
| Total | 272 108 | 12 174 | 2628 | 286 910 |
| Organism . | Number of interactions . | |||
|---|---|---|---|---|
| . | miRNA–mRNA . | ncRNA–protein . | ncRNA–ncRNA . | Total . |
| H. sapiens (human) | 174 853 | 11 026 | 2507 | 188 386 |
| M. musculus (mouse) | 77 270 | 469 | 35 | 77 774 |
| R. norvegicus (rat) | 19 985 | 39 | 1 | 20 025 |
| S. cerevisiae (Baker’s yeast) | 0 | 640 | 85 | 725 |
| Total | 272 108 | 12 174 | 2628 | 286 910 |
The number of miRNA–mRNA, ncRNA–protein and ncRNA–ncRNA interactions per organism in RAIN with a combined confidence score higher than 0.15
| Organism . | Number of interactions . | |||
|---|---|---|---|---|
| . | miRNA–mRNA . | ncRNA–protein . | ncRNA–ncRNA . | Total . |
| H. sapiens (human) | 174 853 | 11 026 | 2507 | 188 386 |
| M. musculus (mouse) | 77 270 | 469 | 35 | 77 774 |
| R. norvegicus (rat) | 19 985 | 39 | 1 | 20 025 |
| S. cerevisiae (Baker’s yeast) | 0 | 640 | 85 | 725 |
| Total | 272 108 | 12 174 | 2628 | 286 910 |
| Organism . | Number of interactions . | |||
|---|---|---|---|---|
| . | miRNA–mRNA . | ncRNA–protein . | ncRNA–ncRNA . | Total . |
| H. sapiens (human) | 174 853 | 11 026 | 2507 | 188 386 |
| M. musculus (mouse) | 77 270 | 469 | 35 | 77 774 |
| R. norvegicus (rat) | 19 985 | 39 | 1 | 20 025 |
| S. cerevisiae (Baker’s yeast) | 0 | 640 | 85 | 725 |
| Total | 272 108 | 12 174 | 2628 | 286 910 |
The human interactions constitute ∼66% of the total RAIN interactions. This likely reflects a research bias towards investigating and annotating ncRNA in human relative to mouse and rat. Saccharomycescerevisiae does not harbor miRNAs and the other constituents of the RNAi pathway, thus miRNA target predictions cannot be provided for this yeast species. The S. cerevisiae genome does, however, encode a wide range of RNA binding proteins and various classes of ncRNA, that have been investigated in the literature, e.g. by pull-down studies (21, 23). Hence, the interactions of several players in transcriptional and post-transcriptional regulation have been integrated and are available in RAIN for all four organisms. We expect that RAIN will be a valuable tool to facilitate the understanding of the molecular regulatory mechanisms. In addition to aiding the researcher in the process of generating hypotheses to be tested, RAIN also allows researcher to advance differential high-throughput studies with a layer of regulatory network biology.
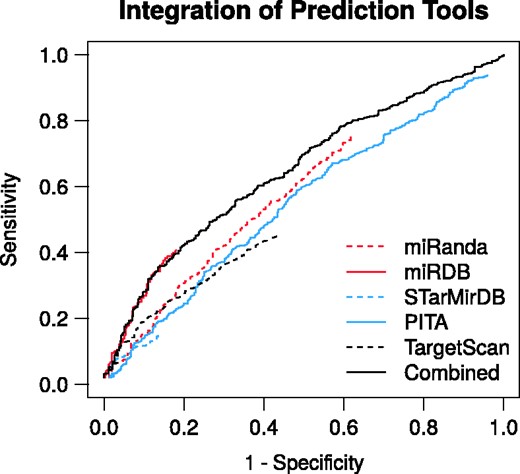
To demonstrate the gain of integrating the individual sources of evidence, we benchmarked the RAIN prediction channel and the respective miRNA target prediction tools. We performed ROC calculations (Figure 3) on the validation set of 1387 positive and 460 negative miRNA–mRNA pairs as described in Section Validation of the integration of miRNA target predictors. The respective miRNA target predictors impose a score threshold for reporting miRNA targets. Hence, despite subjecting all pairs of miRNA and mRNA 3′ UTR to target prediction, only a subset is reported along with a prediction score from each tool (see Supplementary Section 4 for validation set coverage by each tool). Consequently, the ROC curves are truncated as not all positive and negative pairs in the benchmarking sets are reported by the respective prediction tools. This is especially pronounced for tools that rely on conservation of the miRNA target site and 3′ UTR as is the case for TargetScan. The ROC analyses demonstrate that in addition to improving the coverage of the miRNA interactome, integration of the miRNA target predictors also yields an improved predictive performance.
Receiver-operating characteristics of the RAIN prediction channel and the respective miRNA target prediction tools benchmarked against an independent validation set of miRNA–mRNA interactions. The integration of the respective prediction tools yields improved predictive performance. Where specificity , sensitivity , P is the number of positive and n the number of negative miRNA–mRNA pairs.
We restrained the benchmarking of RAIN to the prediction channel, i.e. the integration of miRNA target predictors. The reason is that the publications underlying the validation set are likely overlapping with the literature evidence underlying RAIN text mining, experiments and curated knowledge evidence. The true performance of RAIN is thus underestimated here as it is only based on the weakest of the four evidence channels.
Utility of the Database
This section describes the RAIN website and user interface. An example use case concludes the section and illustrates the utility of the database.
Query interface
Querying RAIN for a single ncRNA or protein identifier returns interactions for this entity; querying for multiple identifiers returns interactions between these entities. After searching RAIN, an identifier disambiguation page allows the user to choose desired query entities among all ncRNA and protein identifiers in RAIN that match the query. RAIN uses STRING v10 (1) protein identifiers for input protein and mRNA identifiers. miRNAs are mapped to miRBase v20 (27) identifiers. For other ncRNAs, RAIN accepts Ensembl (37) and RefSeq (38) identifiers as well as identifiers from four organism-specific main databases obtained from Ensembl BioMart v78 (26): HGNC (34) for human, MGI (35) for mouse, RGD (39) for rat and SGD (36) for yeast.
Network view
After querying RAIN, a search results page featuring a static image of the resulting interaction network is shown. Associations adjacent to ncRNAs obtained from RAIN and protein–protein interactions from STRING are shown in the same network. Sources of evidence supporting an association are indicated by different edge colors. If interactions for a protein were searched, the number of interacting ncRNAs and proteins displayed can be adjusted. Furthermore, the interaction network may be downloaded as files in tab-separated and PSI-MITAB format.
Clicking the network image redirects to an interactive network view in STRING allowing users to adjust the confidence score cutoff and network size as well as centering the network on different nodes. Clicking a node or an edge in the interactive network view lists additional information. For each edge, the confidence score computed for each evidence channel as well as the combined confidence score are shown. Clicking a node shows basic information about the corresponding molecular entity. Information about ncRNAs nodes and adjacent interactions are contributed by RAIN while information about proteins and and protein–protein interaction are provided by STRING.
Data downloads
All interactions and the benchmarking gold standard can be downloaded as tab-separated files from the RAIN website easing programmatic analyses of RAIN data. Interactions are split by evidence channel and, in contrast to those shown in the RAIN network view, not reduced to those interactions with a combined score larger than 0.15. Furthermore, compiled aliases of ncRNA and protein identifiers are available for download.
Use case
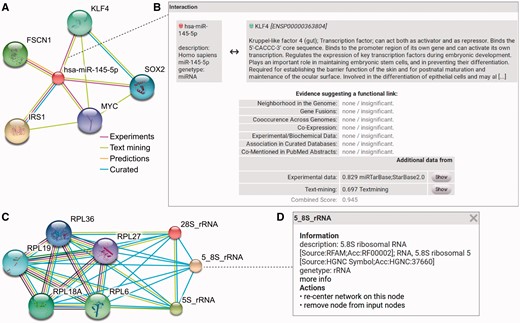
RAIN enables users to study ncRNAs, proteins and their interactions in an intuitive workflow as displayed in Figure 4 and answer complex research questions. Figure 4A exemplifies the single identifier search in RAIN while an example for an edge pop-up window with more information about an association of interest is shown in Figure 4B. Listing the sources of evidence and confidence scores for each interaction make the network easily interpretable. Furthermore, the multiple identifier search in RAIN is shown in Figure 4C. Finally, a node pop-up window with information about ncRNA nodes is depicted in Figure 4D.
RAIN use case. (A) Querying RAIN for human miR-145-5p (miR-145), suggested to act as tumor-suppressor in breast and colon cancer (40, 41), finds multiple oncogenes such as KLF4 and SOX2 (42, 43) as putative targets of miR-145. Evidence channels supporting each interaction are encoded as edge colors. (B) Sources of evidence for each association, e.g. between miR-145 and KLF4, are presented in a pop-up opened after clicking an edge in the network. RAIN confidence scores are collected in the ‘Additional data’ table. Information about KLF4 is provided by STRING. Clicking the ‘Show’ button leads to a website that links to research articles presenting experimental evidence and displaying detailed text mining evidence, where available. (C) In contrast to single identifier search (A), the RAIN multiple identifier search can be used to specifically view interactions between three ribosomal RNAs (28S_rRNA, 5_8S_rRNA, 5S_rRNA) and a subset of five ribosomal proteins part of the large ribosomal subunit. These interactions were extracted from Reactome (10) or found by text mining. (D) Clicking an ncRNA node in the network opens a popup with basic information about the ncRNA, e.g. 5.8S rRNA.
Conclusion
We presented RAIN, a novel database for ncRNAs and their interactions with other ncRNAs and proteins. Associations in RAIN are obtained from a set of resources based on expert curation, experiments, text mining and interaction predictions. RAIN uses a probabilistic scoring scheme to assign a single confidence score to each interaction allowing users to integrate support from all sources of evidence in RAIN in a single number.
RAIN is tightly integrated with the STRING database for protein–protein interactions and adds ncRNAs together with their interactions to the existing protein–protein interaction networks in STRING. RAIN is implemented using the STRING payload mechanism. This allows RAIN users to use interactive and accessible STRING network visualizations. Additionally, potential RAIN users may already be familiar with the STRING interface, further reducing the effort needed to start exploring RAIN.
Future work includes expanding the gold standard to improve the accuracy of the RAIN confidence scores. Furthermore, additional sources of ncRNA interactions such as expert curated interactions from TarBase, which was not included due to current licensing restrictions, could be included. The curated knowledge evidence channel will be expanded to other ncRNA classes and updated according to future literature evidence while maintaining the same high inclusion criteria. The integration of RNA–protein binding site prediction approaches such as RNAcontext (44) or GraphProt (45) would also be of interest. This would, however, require an extension of the gold standard to include this type of interaction. After expanding RAIN to cover more organisms and establishing a comprehensive definition of orthologous groups for ncRNAs, similar to eggNOG (46) for proteins, RAIN interaction evidence could furthermore be transferred between organisms. Finally, further annotations in the node pop-up, e.g. disease association, tissue specificity, and species conservation could prove to be useful. We plan to address these points in future versions of RAIN.
RAIN facilitates the understanding of complex molecular networks through the integration of ncRNA interactions and protein–protein association networks. The graphical web interface provides the researcher with intuitive access to the interactions of ncRNAs and proteins of interest and assigns a confidence score to each association. The incorporation of ncRNAs, including intensely investigated miRNAs and long ncRNAs, makes RAIN a powerful tool to answer current research questions.
Supplementary Data
Supplementary data are available at Database Online.
Funding
The Danish Council for Independent Research (Technology and Production Sciences); The Danish Center for Scientific Computing (DCSC/DEiC); Innovation Fund Denmark (Programme Commission on Strategic Growth Technologies); The Novo Nordisk Foundation (NNF14CC0001).
Conflict of interest. None declared.
References
Author notes
Present address: Christian Garde, The Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Building 6.6 Blegdamsvej 3B, 2200 Copenhagen N, Copenhagen, Denmark
These authors contributed equally to this work.
Citation details: Junge,A., Refsgaard,J.C., Garde,C. et al. RAIN: RNA–protein association and interaction networks. Database (2016) Vol. 2016: article ID baw167; doi:10.1093/database/baw100