-
PDF
- Split View
-
Views
-
Cite
Cite
Zhenzhen Xu, Jing Liu, Wanchao Ni, Zhen Peng, Yue Guo, Wuwei Ye, Fang Huang, Xianggui Zhang, Peng Xu, Qi Guo, Xinlian Shen, Jianchang Du, GrTEdb: the first web-based database of transposable elements in cotton (Gossypium raimondii), Database, Volume 2017, 2017, bax013, https://doi.org/10.1093/database/bax013
Close - Share Icon Share
Abstract
Although several diploid and tetroploid Gossypium species genomes have been sequenced, the well annotated web-based transposable elements (TEs) database is lacking. To better understand the roles of TEs in structural, functional and evolutionary dynamics of the cotton genome, a comprehensive, specific, and user-friendly web-based database, Gossypium raimondii transposable elements database (GrTEdb), was constructed. A total of 14 332 TEs were structurally annotated and clearly categorized in G. raimondii genome, and these elements have been classified into seven distinct superfamilies based on the order of protein-coding domains, structures and/or sequence similarity, including 2929 Copia-like elements, 10 368 Gypsy-like elements, 299 L1, 12 Mutators, 435 PIF-Harbingers, 275 CACTAs and 14 Helitrons. Meanwhile, the web-based sequence browsing, searching, downloading and blast tool were implemented to help users easily and effectively to annotate the TEs or TE fragments in genomic sequences from G. raimondii and other closely related Gossypium species. GrTEdb provides resources and information related with TEs in G. raimondii, and will facilitate gene and genome analyses within or across Gossypium species, evaluating the impact of TEs on their host genomes, and investigating the potential interaction between TEs and protein-coding genes in Gossypium species.
Database URL: http://www.grtedb.org/
Introduction
Transposable elements (TEs) are the most abundant DNA components in most characterized genomes of high eukaryotes (1). Based on their structural features and transposition mechanisms, TEs are generally classified into two classes: retrotransposons and DNA transposons (2). In plants, retrotransposons are further classified into two distinct orders, long terminal repeat (LTR)-retrotransposons (Ty1/Copia and Ty3/Gypsy) and non-LTR retrotransposons (LINE and SINE), whereas DNA transposons are traditionally separated into two main orders, terminal inverted repeat (TIR) (Tc1-Mariner, hAT, Mutator, PIF/Harbinger and CACTA) and Helitron (Helitron) (2, 3). Although TEs are often considered as ‘junk DNA’ due to their continuous reproduction and potential disruption of the regular host genes (4–6), more evidence has unambiguously shown that they play important roles in altering gene structures, regulation of gene expression, affecting genome evolution and creating new genes (7–9). Thus, complete identification and characterization of TEs have become a priority in genome sequencing projects, and this will largely contribute to accurate annotation of protein-coding genes and other genomic components, and play significant roles in investigating potential interaction between TEs and functional genes (10).
Recently, several diploid and tetroploid Gossypium species genomes have been sequenced (11–15), and the availability of their draft genome sequences have provided an unprecedented opportunity for identification, structural and functional characterization and evolutionary analyses of TEs in this economically important crop. Gossypium raimondii (DD; 2n = 6), one of the putative D-genome parent of tetraploid cotton species [such as G. hirsutum (L). and G. barbadense (L.)] has a smaller genome size (∼737.8 Mb) (12). So, we carried out the characterization of almost all families of TEs in G. raimondii genome using comprehensive methods, and constructed the G. raimondii transposable elements database (GrTEdb) in this study. We implemented web-based sequence browsing, searching, downloading and blast tool to help users easily and effectively to annotate the TEs or TE fragments in genomic sequences from G. raimondii and other closely related Gossypium species. Thus, GrTEdb provide the first web-based friendly user interface database of TEs in Gossypium species, and will also facilitate genome evolution analyses within or across Gossypium species, evaluating the impact of TEs on their host genomes, and investigating the potential interaction between TEs and protein-coding genes.
Construction and content of the database
The assembled sequence of the G. raimondii genome was downloaded from http://www.phytozome.com (11). A combination of structure-based and homology-based approaches was employed to identify different TEs in the G. raimondii genome. LTR-retrotransposons were characterized according to the methods previously described by Ma et al. (2006) (16): first, the LTR-retrotransposons were identified using the LTR_STRUC software; then CROSS_MATCH was used to detect elements missed by the program. The alignments were performed between G. raimondii genome and the flanking LTRs of these LTR-retrotransposons, which generated by the LTR_STRUC. Different perl scripts were written to facilitate the data mining and analyses. Other Non-LTR-retrotransposons and DNA transposons (such as L1, Mutator, PIF-Harbinger, CACTA and Helitron) were detected following the protocol provided by Holligan et al. (2006) (17): the alignment were performed between the conservative sequences of transposase in Arabidopsis thaliana and G. raimondii genomes using tblastn, and the TSD and TIR were detected using some perl scripts. The detailed manual inspection was conducted to confirm each predicted element and to define its structure and boundaries. In addition, TEs were classified into different superfamilies and families as previously described (2, 17). Only elements with clearly defined boundaries and insertion sites were deposited in the GrTEdb database.
Based the above approaches, 14 332 TEs were structurally annotated and clearly categorized in the G. raimondii genome, and these elements are classified into seven distinct superfamilies based on the order of protein-coding domains, structures and/or sequence similarity, including 2929 Copia-like elements, 10 368 Gypsy-like elements, 299 L1, 12 Mutators, 435 PIF-Harbingers, 275 CACTAs and 14 Helitrons (Table 1). Based on the 80-80-80 rule (2). TEs that were assigned as Copia- and Gypsy-like elements superfamilies were then categorized into 199 and 218 distinct families respectively because of their large number in G. raimondii.
Summary of the identified TEs in G. raimondii
| Class . | Order . | Superfamily . | Copy numbers . |
|---|---|---|---|
| Retrotransposons | LTR | Copia | 2929 |
| Gypsy | 10 368 | ||
| LINE | L1 | 299 | |
| DNA transposons | TIR | Mutator | 12 |
| PIF-Harbinger | 435 | ||
| CACTA | 275 | ||
| Helitron | Helitron | 14 | |
| Total | 14 332 |
| Class . | Order . | Superfamily . | Copy numbers . |
|---|---|---|---|
| Retrotransposons | LTR | Copia | 2929 |
| Gypsy | 10 368 | ||
| LINE | L1 | 299 | |
| DNA transposons | TIR | Mutator | 12 |
| PIF-Harbinger | 435 | ||
| CACTA | 275 | ||
| Helitron | Helitron | 14 | |
| Total | 14 332 |
Summary of the identified TEs in G. raimondii
| Class . | Order . | Superfamily . | Copy numbers . |
|---|---|---|---|
| Retrotransposons | LTR | Copia | 2929 |
| Gypsy | 10 368 | ||
| LINE | L1 | 299 | |
| DNA transposons | TIR | Mutator | 12 |
| PIF-Harbinger | 435 | ||
| CACTA | 275 | ||
| Helitron | Helitron | 14 | |
| Total | 14 332 |
| Class . | Order . | Superfamily . | Copy numbers . |
|---|---|---|---|
| Retrotransposons | LTR | Copia | 2929 |
| Gypsy | 10 368 | ||
| LINE | L1 | 299 | |
| DNA transposons | TIR | Mutator | 12 |
| PIF-Harbinger | 435 | ||
| CACTA | 275 | ||
| Helitron | Helitron | 14 | |
| Total | 14 332 |
User interface
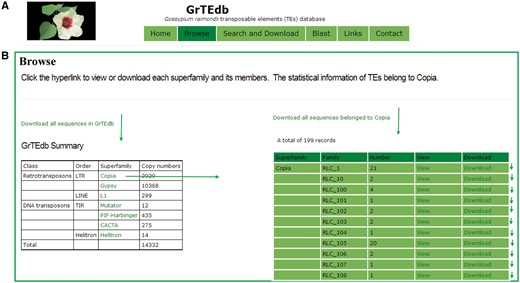
GrTEdb was established to enable users to browse, search, view, analyze and download the TEs data and information. The GrTEdb database organization is navigated by six sections: Home, Browse, Search and Download, Blast, Links and Contact (Figure 1A).
(A) The top menu of GrTEdb. (B) The user interface of browsing in GrTEdb. Users can browse the detailed information of each superfamily by clicking the hyperlinks provided in this page.
Browse
In the browsing interface, the classification structures of TEs deposited in GrTEdb were showed. Users can download the whole TEs sequences, and can browse any one superfamily of interest by the hyperlinks provided. The detailed information of each superfamily can be retrieved and downloaded by clicking the corresponding entry (Figure 1B).
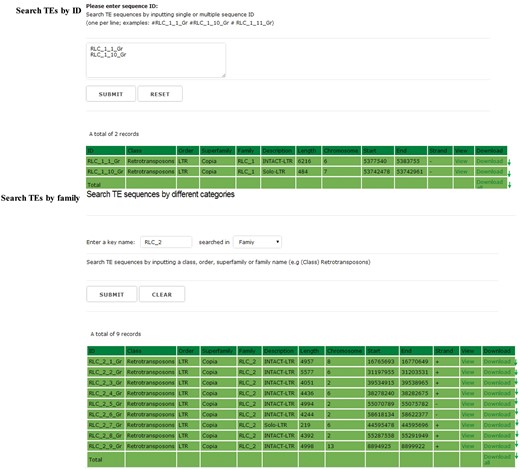
Search and download
In the searching and downloading interface, users can use a keyword to search the GrTEdb (e.g. TE ID, Class, Order, Superfamily and Family) to locate specific TEs quickly. The search results can be viewed and downloaded by clicking the hyperlinks provided on the page (Figure 2).
The searching interface of GrTEdb. Users can use a keyword to locate specific TEs quickly in GrTEdb (e.g. TE ID, Class, Order, Superfamily and Family). The search results can be viewed and downloaded by clicking the hyperlinks provided on the page.
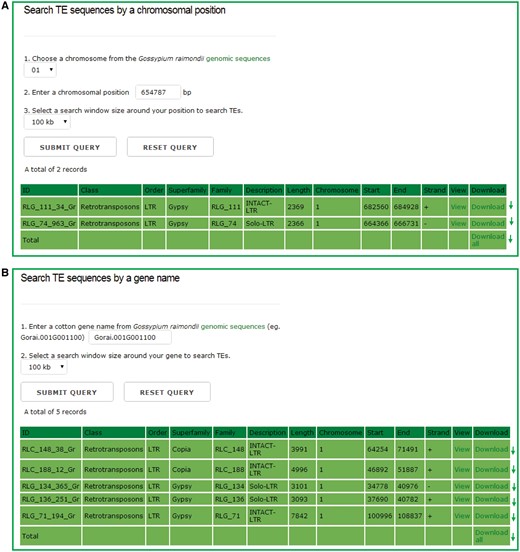
In the chromosomal region search page, users can retrieve the TEs for any one entire chromosome or in a defined window around either a chromosomal position or a gene model, and the detailed information of each retrieved TEs can be viewed and downloaded by clicking the hyperlinks provided on the page (Figure 3). This function can help users to locate TEs that surround the genes of interests easily, and study the interaction between TEs and their adjacent genes.
The chromosomal region search page. Users can retrieve the TE sequences for any one entire chromosome or in a defined window around either a chromosomal position or a gene model, and the detailed information of each retrieved TEs can be viewed and downloaded by clicking the hyperlinks provided on the page.
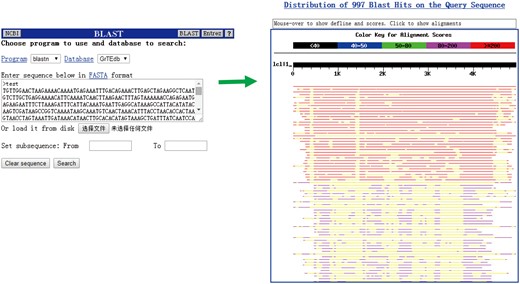
Blast
We did not intend to integrate tools currently available (except for BLAST) for sequence comparison, editing and/or assembly in our database because of the complex structural variation and distribution patterns of TEs among classes and families (Figure 4). In the BLAST search page, users can handy and quickly compare their sequences with the cotton TEs deposited in GrTEdb.
The BLAST interface (left) and a sample of BLASTn results (right) provided in GrTEdb.
Links
A variety of links to other TEs database were included in our GrTEdb database.
Contact
In this section, contact information and links to our labs were provided. Please feel free to contact us if you have any suggestions and problems.
Discussion
Because of the structural complexity and the time consuming process, it remains challenging to annotate all TEs in a sequenced genome. Currently only a few TE databases have been established (10, 18–24). Because these databases can help users easily and quickly annotate their sequences, and they have been widely used (10). However, in these plant TE databases such as P-MITE (a Plant MITE database), the TIGR Plant Repeat Databases, and so on, there is little information about the cotton TEs. In parallel, although there were some reports associated with TEs in Gossypium (11–15, 25, 26), the web-based database of TEs was lacked. Here we have generated a web-based TE database (GrTEdb) using multiple methods, and only TEs with clearly defined boundaries were deposited in the database. More studies have showed that many TEs are structurally incomplete because they have undergone intra- or inter-element unequal recombination or accumulation of small deletions by illegitimate recombination (27, 28). For example, a large number of LTR-RT families with highly degraded protein-coding sequences or without any coding sequences (often defined as non-autonomous elements) have been found in several plants (29–35), and these elements remains challenging to be identified and characterized. Therefore, GrTEdb provides the reference sequences of TEs data for cotton, and users can use these data to identify more complex elements and develop their specific functions.
Recently, G. arboretum (A2) genome, a pupative contributor of the A subgenomes cotton species, and the allotetraploid upland cotton (AD)1 [G. hirsutum (L.)], which accounts for >90% of cultivated cotton worldwide, have been sequenced and assembled (13–15). Because of the close evolutionary relationships of DD, AA and AADD genomes, our GrTEdb database is not only useful for G. raimondii study, but also can facilitate structural and evolutionary analysis in AA, DD, AADD and other unfinished Gossypium genomes. The web-based interface can also help users at the beginning stage of bioinformatics to easily access and use this database. Further, TEs in our database will help cotton breeders develop markers for mapping agronomically important genes and accelerate breeding process.
Conclusions
We have generated a web-based GrTEdb, and it provides researchers with not only resources and information related to different TEs in the cotton genome but also tools for performing data analysis. Thus, GrTEdb will facilitate cotton genome evolution analyses among AA, DD and AADD genome species, the evaluating impact of TEs on their host genomes, and investigating the potential interaction between TEs and protein-coding genes. In parallel, TEs in our database will facilitate users for marker development for mapping agronomically important genes, and for both intra- and inter-specific comparison of TEs at whole genome levels.
Availability and requirements
All TEs or subsets of TEs can be viewed and downloaded from the website http://www.grtedb.org/, and all data deposited in the database are freely available to all users without any restrictions.
Funding
The Key Scientific and Technological Project of Jiangsu Province (BK20150540); Jiangsu Agricultural Science and Technology Innovation Fund (CX(14)5008); the State Key Laboratory of Cotton Biology Open Fund (CB2016B03); the National Natural Science Foundation of China (NSFC) (31370266, 31471545).
Conflict of interest. None declared.
References
Author notes
These authors contributed equally to this work.
Citation details: Xu,Z., Liu,J., Ni,W. et al. GrTEdb: the first web-based database of transposable elements in cotton (Gossypium raimondii). Database (2017) Vol. 2017: article ID bax013; doi:10.1093/database/bax013