-
PDF
- Split View
-
Views
-
Cite
Cite
Olivier Arnaiz, Agata Malinowska, Catherine Klotz, Linda Sperling, Michal Dadlez, France Koll, Jean Cohen, Cildb: a knowledgebase for centrosomes and cilia, Database, Volume 2009, 2009, bap022, https://doi.org/10.1093/database/bap022
Close - Share Icon Share
Abstract
Ciliopathies, pleiotropic diseases provoked by defects in the structure or function of cilia or flagella, reflect the multiple roles of cilia during development, in stem cells, in somatic organs and germ cells. High throughput studies have revealed several hundred proteins that are involved in the composition, function or biogenesis of cilia. The corresponding genes are potential candidates for orphan ciliopathies. To study ciliary genes, model organisms are used in which particular questions on motility, sensory or developmental functions can be approached by genetics. In the course of high throughput studies of cilia in Paramecium tetraurelia, we were confronted with the problem of comparing our results with those obtained in other model organisms. We therefore developed a novel knowledgebase, Cildb, that integrates ciliary data from heterogeneous sources. Cildb links orthology relationships among 18 species to high throughput ciliary studies, and to OMIM data on human hereditary diseases. The web interface of Cildb comprises three tools, BioMart for complex queries, BLAST for sequence homology searches and GBrowse for browsing the human genome in relation to OMIM information for human diseases. Cildb can be used for interspecies comparisons, building candidate ciliary proteomes in any species, or identifying candidate ciliopathy genes.
Database URL:http://cildb.cgm.cnrs-gif.fr
Introduction
Ciliopathies represent a class of genetic diseases attributed to dysfunction of centrioles and their associated structures and/or derivatives, centrosomes and cilia (1–3). Indeed, the centriole, a barrel-shaped cylinder of triplets of microtubules, fulfills a wide range of functions that can be classified into two broad categories. First, when in the cytoplasm, a complex matrix of proteins is assembled around the centrioles, thus forming the centrosome, a microtubule organizing platform directly involved in cell shape, cell polarity and cell division. Second, when anchored in the plasma membrane, the centriole behaves as a basal body and nucleates an axoneme, which, depending on species and/or cell type, can be the backbone of flagella or cilia, be they motile or immotile, sensory or primary cilia (4–6). Recent high throughput studies revealed that these structures are composed of hundreds of proteins. Presently known ciliopathies such as Kartagener, Bardet-Biedl, Meckel-Grüber, Alstrom, Joubert syndromes, polycystic kidney disease, originate from ciliary dysfunction during embryonic development and in adult organs, and provoke a combination of multiple symptoms in patients, such as polydactyly, obesity, sterility, mental retardation, kidney polycystosis, deafness, retinal defects, ciliary dyskinesia, sinusitis, otitis, bronchiectasis (7). From the high complexity in protein composition of centriole/basal bodies and cilia/flagella, we can anticipate a growing number of known and orphan diseases to be identified as ciliopathies.
Cilia can have motile functions and are involved in fluid movement (e.g. mucous, cerebral-spinal fluid). They can also have sensory functions (e.g. in olfactory neurons, photoreceptors). Primary cilia play prominent roles in development (8) and most likely also in tissue maintenance and regeneration since they are present on stem cells (9). The ciliary axoneme is a cytoskeletal structure highly conserved through evolution, formed by a cylinder of nine doublets of microtubules plus, in most cases, a central pair of microtubules (9 + 2 pattern), enveloped by an extension of the plasma membrane. The ciliary membrane contains ion channels, receptors and other signaling proteins that control axoneme bending for motility or that sense chemical or mechanical stimuli and transduce signals internally (6). Not only is the ultrastructure conserved, but also the core protein composition, so that most protein functions can be extrapolated from one species to another, hence the development of model systems for ciliary function, such as Caenorhabditis, Chlamydomonas, Drosophila, Paramecium, Tetrahymena and Trypanosoma.
Our laboratory, taking advantage of the Paramecium genome sequence (10) and the ParameciumDB model organism database (11), focuses on motile and sensory aspects of ciliary function in Paramecium. Advantages of the model include rapid and efficient RNAi and easy phenotypic description of swimming behavior that relies on ciliary function. We performed high throughput analyses on Paramecium cilia: a proteomics study of isolated cilia (see Supplemental Data) and a study of transcriptome changes during ciliary biogenesis (Arnaiz et al., in preparation). We were confronted with the problem of comparing our data with previous studies of centrioles and cilia in other organisms. This prompted us to build Cildb (http://cildb.cgm.cnrs-gif.fr), a knowledgebase that relates whole proteomes of 18 species through orthology and links the relevant proteins to ciliary studies as well as to the OMIM database of human genetic diseases. Cildb was designed for finding information on cilia and ciliopathies, as illustrated here. In addition, as it contains the whole proteome of each species, Cildb has wider applications such as linking any genetic disease to model organisms.
Objectives and specifications
Three databases related to centrosome, basal bodies and cilia/flagella are currently available, the Centrosomedb [(12), http://centrosome.dacya.ucm.es/], the Ciliome Database [(13), http://www.sfu.ca/∼leroux/ciliome_database.htm] and the Ciliaproteome [(14), http://v3.ciliaproteome.org/cgi-bin/index.php]. However, none of these databases met our needs since the genome data on Paramecium is not incorporated, no entry to these databases is possible using sequence information, the raw data from each original study is not available, and the orthology relationships between species were calculated only by BLAST best reciprocal hits, which masks multigene families. Altogether, this led us to design a completely new tool to browse the complex data emanating from very different approaches (proteomics, transcriptomics, comparative genomics, search for promoters) in different model organisms.
A database dedicated to studies of an organelle, such as centriole or cilium, must be open to future discoveries and identification of novel proteins not yet found in present studies. It is thus necessary for the the whole proteome of each species to be present in the database. Comparing studies made in different species implies that orthology relationships have to be calculated between species, using an algorithm that takes account of multigene families (a particularly important consideration for the Paramecium genome, but extant in all genomes). In addition, it seemed worthwhile to allow queries using any species as the entry proteome. This implies that orthology calculations have to be made for each pairwise combination of species in the database. To link the studies to the proteomes, the best would be to incorporate raw data and use them to assign confidence stringency values to the ciliary proteins identified in queries. It also seemed useful to include data from the OMIM database, not only data that identifies genes responsible for human diseases, but also information on diseases imprecisely mapped on the genome and that have many candidate genes in the disease interval. Finally, it seemed that access to the data in Cildb should be versatile and include not only complex queries but also sequence homology searches and a human genome browser that incorporates tracks with ciliary data and OMIM information for navigation along chromosomal regions.
Data sources and computational analyses
Species whose whole proteome is included in Cildb
The present Cildb version, which will be updated at regular intervals in the future, contains the complete set of predicted proteins from the genomes of 18 species, nine of them chosen because high throughput studies on cilia, flagella or centrosomes are available (Caenorhabditis elegans, Chlamydomonas reinhardtii, Drosophila melanogaster, Homo sapiens, Mus musculus, Paramecium tetraurelia, Rattus norvegicus, Trypanosoma brucei, Tetrahymena thermophila), four of them because the organisms are good models for ciliary experiments although no high throughput study is yet published (Ciona intestinalis, Danio rerio, Giardia lamblia, Plasmodium falciparum) and five of them because they lack cilia and centrioles (Arabidopsis thaliana, Dictyostelium discoideum, Escherichia coli, Saccharomyces cerevisiae, Schizosaccharomyces pombe) and can be used in genomic subtractive studies. Sequence information was retrieved either from the EnsEMBL portal when available, or from the database dedicated to the organism (Supplementary Table S1).
Ciliary studies included in Cildb
The ciliary studies incorporated in this version of Cildb are proteomics of centrosome, basal bodies and cilia/flagella of Chalmydomonas, Homo, Rattus, Paramecium, Tetrahymena and Trypanosoma, trancriptome analyses related to the presence of cilia in certain tissues or to ciliary biogenesis in Caenorhabditis, Chlamydomonas and Paramecium, comparative genomics between Homo, Chlamydomonas and Arabidopsis, and search for motifs in promoters in Caenorhabditis and Drosophila (Figure 1, Supplementary Table S1).
Orthology calculations and links to ciliary studies underlying Cildb. The 18 species analyzed are represented as circle arcs proportional to the size of the proteome, in red when ciliary studies exist (delineated as ticks outside the circle), in orange when centriole/cilia exist, but without high throughput studies, and in blue when no cilia or centriole exists (At: Arabidopsis thaliana; Ce: Caenorhabditis elegans; Ci: Ciona intestinalis; Cr: Chlamydomonas reinhardtii; Dd: Dictyostelium discoideum; Dm: Drosophila melanogaster; Dr: Danio rerio; Ec: Escherichia coli; Gl: Giardia lamblia; Hs: Homo sapiens; Mm: Mus musculus; Pf: Plasmodium falciparum; Pt: Paramecium tetraurelia; Rn: Rattus norvegicus; Sc: Saccharomyces cerevisiae; Sp: Schizosaccharomyces pombe; Tb: Trypanosoma brucei; Tt: Tetrahymena thermophila). The colors of the ticks for the studies are explained in the inset to distinguish proteomic, transcriptomic and comparative genomic studies. Andersen (35); Arnaiz [this article for proteomics, or Arnaiz et al. (in preparation) for transcriptome analysis]; Avidor-Reiss (36); Blacque (37); Broadhead (38); Chen (39); Efimenko (40); Keller (41); Kilburn (42); Laurençon (43); Li (16); Liu (44); Mayer (45); Ostrowski (19); Pazour (26); Smith (18); Stolc (46). For each species, the whole proteome was compared by BLASTp to itself and to all the other ones, in order to apply our three orthology filters (Inparanoid, Inparanoid + filtered best hits, BLASTp cutoff), as described in experimental procedures. The high throughput ciliary studies were then remapped to the proteins, so that links between orthology and ciliary properties are created. The genome versions and the origin of the ciliary study data in Cildb are reported in Supplementary Table S1. The human genome is also linked to the OMIM database in which genetic disorders appear.
Determination of orthology relationships
Since whole proteomes may contain splicing variants as well as sets of similar paralogs in multigene families, the way to establish orthology is not straightforward: proteins often cannot be involved in binary orthology relationships identified by best reciprocal BLASTp hits. The Inparanoid program (15) overcomes this problem by relating groups of proteins in one species to groups of proteins in another one, provided that the intraspecific distances between paralogs are shorter than the interspecific distances between the putative orthologs. We thus calculated orthology between the predicted proteins of the 18 genomes cited above using Inparanoid on the output of the pairwise BLASTp comparison between the 18 proteomes, including self-comparisons, 324 comparisons altogether (Figure 1), with the default Inparanoid parameters for eukaryotes.
The stringency of Inparanoid is such that, in case of poor gene annotation (as currently occurs in genome projects with gene truncation, false splicing pattern, gene fusion or splitting), some orthology relationships are missed, leading to an excess of false negatives when given proteins are searched for through orthology in another species. Hence, we added to this calculation the result of analysis of alignments using empirical filters that were manually validated. We carried out Smith–Waterman alignments of all pairs of best hits by BLASTp and added to the Inparanoid results the alignments with at least 30% identity on at least 52% of the length of both proteins, or when the product of both parameters was above 2300 (tolerating better alignments over shorter lengths or the converse).
Determination of homologs for genome subtraction
For some purposes, both of the above methods are too stringent: if they allow recognition of possible orthologs in two proteomes, they cannot prove that a given protein has no ortholog in a species. Comparative genomics often relies on the presence or absence of sets of orthologs in several species. In such cases, less stringent comparison tools are used and a cutoff of 1e − 10 (16) or even higher (17) is employed. We thus also performed a third, low stringency, calculation with a simple cutoff on the BLAST score. Indeed, the BLAST score is much more consistent than the e-value when comparisons are made between proteomes of very different size. The threshold for homolog detection was empirically fixed to ≥70, a score value generally corresponding to an e-value around 1e−10.
Remapping ciliary studies
To identify centrosome, centriole, basal body and flagellar/ciliary proteins, we remapped all studies published to date on the whole proteomes of the corresponding species. This was performed in two steps, retrieving the protein sequences from the original studies and determining the correspondence of these proteins with the recent proteome versions used for orthology calculations.
We started with 21 studies published in 17 articles, including this one, and concerning nine species altogether (Figure 1; Supplementary Table S1). In most of the articles, supplemental tables give the list of protein accession numbers, which permitted retrieval of the sequences (except for a small minority for which the accession number did not correspond to anything). Two studies provided a list of peptides obtained in proteomics, instead of (18) or in addition to (19) a list of protein accession numbers. In such cases, we mapped the peptides to the present version of the proteomes and retrieved the corresponding proteins. Each protein recovered from the ciliary studies has been flagged with attributes corresponding to the raw results in the studies when available (number of peptides in proteomics, fold change or false discovery rate in transcriptome analyses, score and distance from ATG of X-boxes, e-values in comparative genomics). Altogether, 16 038 protein-study entries were recovered (Supplementary Table S1).
When the genome version used in any given study was different from the version we used for orthology determination, we remapped the proteins to the version used for orthology determination using BLASTp. Indeed, from version to version, the genome annotation evolves, some gene models appear while others disappear, and the structure of some others is modified leading to significant changes in the corresponding protein sequence. We considered a protein as remapped if there was at least 90% identity on 90% of the length of each protein. The remaining proteins were validated or rejected by visual examination of the alignments. In this process, 288 entries were dropped because of no hit in the new genome versions, and 243 rejected by human curation. The 15 443 remaining proteins correspond to 19 051 entries in the new genome versions (the discrepancy between the numbers arises from different treatment of alternative splicing, of paralog families, etc. in the various genome versions). The vast majority of the entries (17 348) were automatically remapped, while 1703 of them were recovered by human curation (Supplementary Table S1).
In addition to the protein ‘flagging’ with ciliary studies, we linked all proteins of the proteomes to general attributes predicted from the amino acid sequence such as molecular weight, isoelectric point, presence of signal peptides (20) and number of transmembrane helices (21).
OMIM data integration
The OMIM database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim) gathers all information about genetic inheritance in man, linking genetic diseases to their corresponding genes when they are known. In addition, genetic diseases still imprecisely localized to one or several chromosome bands are also included in the OMIM database, so that candidate genes encompassed by the chromosomal region can be extracted. In Cildb, we used this information in two different ways. First, we flagged human proteins with the OMIM entry when the direct link exists. The OMIM information is then treated like the other properties. Second, when OMIM entries correspond to several genes, we incorporate the information into a distinct database. In this section of Cildb, there are however as many entries as pairs of candidate protein-disease (530 765 altogether, a number much greater than both 46 591, the number of human proteins, and 11 152, the number of OMIM entries referenced in Cildb). Searches can be done to reveal all human genes present within the genetic region where the disease has been genetically localized, and to display them with any desired attribute, including orthology and occurrence in ciliary studies.
Cildb ARCHITECTURE
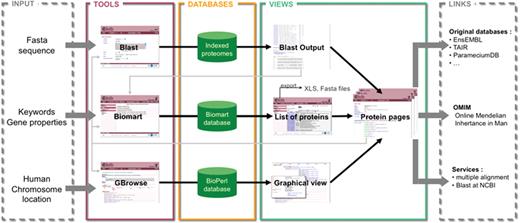
The Cildb web site is organized around three main tools, BioMart, NCBI BLAST and GBrowse (Figure 2). Each tool provides its own interface and its own storage system.
Schema of the structure and possibilities of use of Cildb. The orthology calculations and links to ciliary studies and to OMIM are at the center of Cildb. To access the data, three possibilities of queries are offered: BioMart query using key words or properties of proteins (orthology, ciliary studies, etc.); BLAST of a sequence; browsing human chromosomes with GBrowse. The BioMart (22) tool of Cildb allows the user to build a complex query using a system of filters and to display the information, pre-calculated in the database (PostgreSQL mart database). The result is a list of proteins matching the different criteria. Each protein in the list is linked to a ‘Protein page’, which describes all the information related to this protein. Data in the table can be exported as xls or tsv files, but also the corresponding sequences as fasta files. The BLAST tool uses a search by sequence alignment with an NCBI BLAST interface (23), regular BLAST, PSI-BLAST and PHI-BLAST can be performed. The user can select proteins in the BLAST output and analyze them with BioMart or go to their protein pages. The GBrowse tool (MySQL Bio::DB::SeqFeature::Store database) allows navigation through the human chromosomes to see the genes and proteins with their links to orthologs, ciliary studies and OMIM entries. The user can also analyze the proteins with BioMart or go to the protein page. The protein page itself contains a summary of all the information contained in Cildb for the protein, with internal links to the orthologs, to a BioMart interface, to Cildb BLAST links and to GBrowse for human proteins, and external links to the genuine databases for the accession ID, to OMIM entries, to BLAST at NCBI and to multiple alignment servers.
BioMart is a data management system that provides a powerful complex web query interface (22). Programmatic execution of queries is also available via a web-services API, or direct-access software libraries written in Perl. The data are split into three databases (PostgreSQL) corresponding to the three levels of confidence of the orthology. The databases contain 18 datasets (or marts) according to the 18 species. A dataset is a collection of several tables, which follow a BioMart naming convention (‘dataset__content__type’).
The NCBI BLAST tool (regular, PSI or PHI BLAST) allows homology searches starting from a protein or DNA sequence (23). We have indexed the proteome of each species separately so that it is possible to query any given proteome or all of the proteomes in Cildb taken together.
The Generic Genome Browser (GBrowse version 1.69), the most popular viewer in the GMOD project (24), composed of a web interface (CGI in Perl) and a BioPerl database (Bio::DB::SeqFeature::Store in MySQL), has been implemented in Cildb for the human genome. We generate GFF3 files (Generic File Format version 3) corresponding to each track: human proteins, Inparanoid orthologs and OMIM entries. The OMIM track uses the glyph ‘wave’ (Bio::Graphics::Glyph::wave).
In addition, Cildb contains protein pages, which gather all the information stored in Cildb for any protein. These pages are built using a Model View Controller (MVC) system implemented in Perl with the Template::Toolkit module (25).
Screenshot of a typical query page of Cildb, here using the Homo sapiens whole proteome as a dataset and displaying the categories of filters that can be used for the query.
Cildb UPDATES
Updates of Cildb will be performed periodically after curation of the literature to incorporate new ciliary studies and new proteomes from the corresponding species. We also plan to incorporate new proteomes of species whose phylogenetic position is of interest, whether or not ciliary studies are available. Genomes already in Cildb will also be updated periodically. This means that Inparanoid calculations must be carried out for major genome releases with concomitant remapping of ciliary studies to the proteomes. This procedure is CPU-intensive and also requires human curation, so that we plan to make such updates every 18 months. The next version of Cildb (V2.0) is in early steps of preparation.
User Interface
Cildb can be entered from any of its 18 species and gives access to properties identified in any genome through orthology relationships, whatever the species users are interested in. The three tools described above, BioMart, BLAST and GBrowse are available. We will present the use of these tools in Cildb.
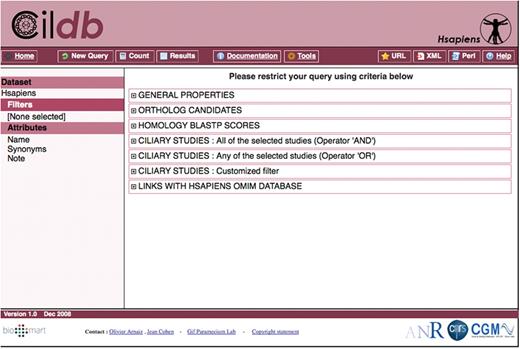
A complex BioMart query is decomposed into four steps: choice of the dataset, filtering of the dataset to select only the proteins with the desired properties, choice of the attributes to be displayed with the output, which will usually be different from the properties used for the query and data retrieval. For simple key word queries, we added a quick search box accessible from every Cildb page.
Datasets: To choose the data set, two operations are needed. First, the user has to choose the orthology/homology calculation method, Inparanoid, Inparanoid plus filtered best hits, Filtered BLASTp, according to the desired use of the database. Then the reference species, among the 18 that are listed has to be chosen.
Filters: Filtration is made using the properties and makes it possible to retrieve only proteins with the desired properties. On the BioMart page (Figure 3), they are grouped in six categories. Attributes: The choice of attributes determines the properties that are displayed for each protein retrieved by the query. The attributes are organized by species. In the reference species (the one chosen as dataset at the beginning of the query, listed on the first line), numerous fields are found: protein ID, synonyms, description, molecular weight, isoelectric pH, presence of transmembrane helices, signal peptides, etc. Since we linked human proteins to the OMIM entries for human genetic disease, if a protein of a given species has a human ortholog referenced in OMIM, this can be displayed as an attribute in the output of the query. When ciliary studies have been conducted in this species, the detailed raw results of the studies are given for each relevant protein. The attributes concerning other species reflect the presence or not of orthologs (according to the method originally selected in the dataset) and, when they exist, whether they have been found in ciliary studies in this species. Detailed results of ciliary studies are not provided for orthologs since a given protein may have several orthologs (i.e. in-paralogs in the Inparanoid family), generating different sets of raw results that cannot be displayed in the BioMart interface. For simpler searching and posting, we also classified the raw results of ciliary studies as low, medium and high stringency, described in detail in Supplementary Table S1. For example, in proteomic analyses, low stringency means identification of a protein by one peptide detected by mass spectrometry, medium stringency by at least two peptides, high stringency by at least four peptides.
– The general filter is used to look for proteins according to general properties, ID number, synonyms, key words in the reference species or in linked orthology groups, molecular weight, isoelectric pH, etc.
– The orthology filter permits the user to select proteins with (or without) orthologs in any combination of species.
– There are three ciliary filters to look for proteins identified in ciliary studies or whose orthologs are identified in ciliary studies. The three filters are different in that the advanced ciliary study filter examines raw results of ciliary studies, the ciliary study filter (all) examines ciliary results according to pre-calculated stringency with multiple selection using the Boolean operator ‘AND’, while the ciliary study filter (any) uses the Boolean operator ‘OR’.
– The OMIM filter permits selection of proteins whose human orthologs are indexed in OMIM.
Whenever sequence information is needed for the output of a query, the ‘sequence’ button can be selected from the attribute page.
Data retrieval: The ‘count’ button displays the number of proteins that pass the filter out of the total number of proteins in the proteome. The ‘results’ button gives access to the list of matching proteins, displayed with all selected attributes. Navigation to the filter or attribute pages makes it easy to refine the search or the display. The results may be exported as text or xls files (or in fasta format for sequences) for further analyses. Cildb results as well as xls exports contain internal links to protein pages in which a summary of all Cildb information on the protein is displayed, as well as internal and external links (Figure 2).
The second way to enter Cildb is via the BLAST tool. This allows the user to retrieve proteins from Cildb, either in the whole database or for a given species, by sequence alignment using the NCBI BLAST algorithm. When performed on a single organism, the Cildb BLAST output provides, in addition to classical alignment output, links to protein pages of Cildb as well as to BioMart views of target proteins, to be filtered by other criteria or displayed with Cildb attributes.
Finally, Cildb can enter the human genome through GBrowse. In addition to tracks to visualize genes and encoded proteins, browsing the human genome in Cildb displays tracks for two kinds of OMIM data, the OMIM description of the corresponding gene and associated diseases if they exist and the overall localizations on the chromosomes of OMIM entries not precisely allocated to a gene, but rather to a chromosomal region. Whether a given protein has been identified through ciliary studies (either directly or through one of its orthologs), or has Inparanoid orthologs, is indicated as a track in the browser.
Use CASES
The applications of Cildb are limited only by the needs and the imagination of the user. First of all, just listing proteins found in particular studies with many sorts of attributes is already a powerful improvement in the field. However, the major innovation of this database consists in allowing all kinds of experiments to look for particular proteins with defined properties in any combination, be they biophysical, from descriptors, orthology relationships, ciliary studies, or in relation to OMIM information.
Comparison of Ciliary proteomics in Paramecium to other ciliary studies
Our work on ciliary proteomics in Paramecium, presented in the Supplementary Data (Supplementary Figure S1, Supplementary Tables S2 and S3), was evaluated by comparison to other ciliary studies performed formerly in other species. This evaluation was performed using the BioMart query tool of Cildb (Figure 4). The figure clearly shows that: (i) in each species, approximately half of the ciliary proteins identified through ciliary proteomics possess orthologs in the other species. Only Paramecium and Tetrahymena, closer in evolution than the other species, share more orthologs. (ii) Our Paramecium ciliary proteomics is more specific than the other ones in the comparison, although less sensitive than the one of Pazour and colleagues (26) in Chlamydomonas. Indeed, no ribosomal or histone contaminants were detected in Paramecium cilia preparations. On the other hand, starting from the ciliary proteome in Paramecium, Tetrahymena or Trypanosoma, a high proportion of the corresponding Chlamydomonas orthologs were already identified by the Chlamydomonas study (26) (dark green in the histograms), whereas the orthologs of Chlamydomonas ciliary proteins identified in ref. (26) were identified in the ciliary proteomics in each other species at a maximal rate of 50%. Pairwise comparisons of ciliary proteomic studies show that our Paramecium ciliary study ranks just after the study in Chlamydomonas in terms of sensitivity, probably owing to the fact that only whole purified cilia were analyzed in Paramecium, in contrast to Chlamydomonas where sub-fractions of cilia were also analyzed by mass spectrometry.
Comparison of ciliary proteomic studies from different unicellular models. Using Cildb, we compared the proteomic studies of purified cilia/flagella of Chalmydomonas, Paramecium, Tetrahymena and Trypanosoma. The protocol was the same for each study: (i) Build a BioMart ‘new query’ in Cildb using the ‘Inparanoid and filtered best hits’ as homology method. (ii) Choose the species in which the ciliary study examined was conducted (e.g. Chlamydomonas). (iii) Filter proteins identified in the relevant proteomic study (e.g. Pazour’s proteomics) with medium stringency confidence (two or more different peptides). (iv) Select the ‘number of ciliary studies in all species with medium confidence’ as an attribute. (v) Count and display the results as xls tables. In the first column of each graph, the total height represents the number of proteins identified in the study, the dark blue proteins are those found as ciliary in at least another study, pale blue proteins are those found as ciliary only in this study, and grey proteins are those annotated as ribosomal proteins or histones, representing likely contaminants of the ciliary preparation. (vi) Filter proteins using the same criteria as in (iii) as well as the existence of ‘orthologs’ in each species used in this comparison (the other protists plus human). This represents as many queries as species examined and gives the height of the green bars for each species (the grey part corresponding to ribosomal proteins and histones). (vii) Filter as in (vi) and look in the results for proteins identified by ciliary proteomics in the target species [e.g. this study for Paramecium or Broadhead et al. (38) for Trypanosoma], represented as dark green, whether they have been found as ciliary in other species (medium green) or not (pale green).
Building a new ciliary proteome
Using Cildb, one can identify proteins likely to be constituents of cilia even in species devoid of any specific ciliary study, e.g. Danio rerio, Giardia lamblia or Ciona intestinalis, just by looking in this species for proteins having orthologs in other species that have been identified as ciliary proteins. Supplementary Table S4 gives an example of a Cildb output in xls format, in this case a list of the 975 proteins of Danio rerio with orthologs in at least three different species identified as a ciliary protein with medium confidence (see definition in Supplementary Table S1).
Real-time comparative genomics
Comparative genomics provided a powerful strategy for the identification of centriole/cilia proteins by considering all Chlamydomonas reinhardtii proteins having an ortholog in Homo sapiens but not in Arabidopsis thaliana (subtraction of ‘non-ciliary genomes’ from the common protein complement between two ‘ciliary genomes’) (16). Similar experiments can now be done online using Cildb, with any combination of species. For that purpose, we provide homology based on a BLAST score cutoff of 70 (see Experimental Procedures section in Supplementary Data), as did the above-mentioned study with a 1e−10 cutoff (16), rather than on Inparanoid orthology, to avoid an abundance of false negatives. The exact reproduction of the experiment reported in ref. (16), using Cildb is presented in Supplementary Tables S5a and S5b. The interest here is that the in silico experiment using Cildb can be validated by ‘bench’ experiments compiled in the database, just by looking at the experimental ciliary attributes of the identified proteins. Despite good overall concordance, differences appear between the original experiment and the Cildb screen, but careful examination reveals that all differences arise from the evolution of the annotation between successive genome versions: genes found with the Cildb screen but not by Li et al. (16), correspond to gene models present in version 3 but not in version 2 of the Chlamydomonas genome; conversely, genes not found with Cildb orthology but identified by Li et al. (16), all correspond to gene models present in the recent version of the Arabidopsis genome but not in the former one (so that these ‘non-ciliary’ proteins could not be subtracted from the data set at that time). Thus, the procedure used in Cildb online supports comparative genomics experiments using any combination of the 18 species currently available.
Identification of ciliopathy genes
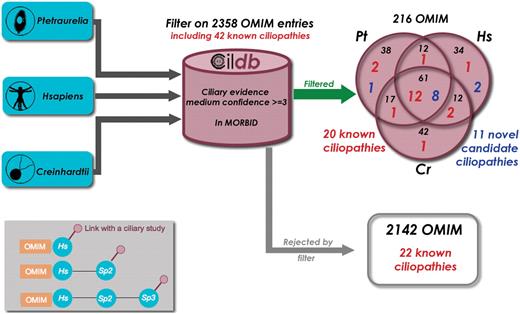
From global analyses that can be mined in human or model organisms using Cildb, at least a thousand proteins are likely to be components of, or involved in the biogenesis of centrioles, basal bodies, cilia and flagella. Dysfunction of some of these proteins leads to severe diseases, the ciliopathies. Recent reviews (27,28) listed known ciliopathies and a few more can be retrieved from the literature: CILD6 due to mutations in TXNDC3 (29), CILD9 to mutations in DNAI2 (30), CILD10 to mutations in KTU (31), and CIL11 and CIL12 to mutations in RSPH4a and RSPH9, respectively (32). Altogether, 50 human genes are involved in ciliopathies when they are mutated. Eight of these cannot yet be retrieved from Cildb, because the version of OMIM incorporated in Cildb does not yet display the links to these disorders. To assess the interest of Cildb to find ciliopathies, we performed a BioMart query starting successively from H. sapiens, C. reinhardtii and P. tetraurelia, filtering proteins for their links to a human disorder (2358 entries in the MORBID section of OMIM) and requiring at least three ciliary studies with a medium confidence stringency (Figure 5). Proteins linked to 216 disorders appear in the filtered output, including 20 of the 42 indexed ciliopathies. This represents a ciliopathy enrichment by a factor of 5 (compare 20 out of 216 with 42 out of 2358). In addition, the ciliopathy CIL6, not listed in the reviews, appears in this search. This may indicate that many of the 216 disorders identified in this query may be novel ciliopathy-related diseases. The 22 ciliopathies not found may be explained by the fact that not all ciliary proteins are identified in high throughput studies.
Finding novel ciliopathy candidate genes with Cildb. Starting successively with the proteomes of three species, Paramecium tetraurelia, Homo sapiens and Chlamydomonas reinhardtii, we applied the same BioMart filters, ‘at least 3 ciliary evidences with medium confidence’ and ‘has a human ortholog linked to a disorder reported in OMIM’. As a result, we extracted the OMIM entries linked to a human disorder, as revealed by the filter applied to each species. Of the 2358 disorders present in the GeneMap section of OMIM, 216 passed the filter and 20 of the 42 known ciliopathies with links with a disorder entry in OMIM (Table 1) were found. The 2142 other disorders include the 22 remaining known ciliopathies. This is not surprising since many ciliopathy genes are not revealed by high throughput studies. Nevertheless ciliopathy genes are highly enriched by our filter (1/10 versus 1/100). Detailed examination of the other disorders provided by the filter allowed us to propose 11 of them as candidate ciliopathies (see Table 1). Inset: the filter used results in linking OMIM entries to ciliary studies, however by passing through their association with a human gene. Three configurations can provide these links. (i) the human protein is directly concerned by a high throughput study in man. (ii) the human protein displays a ciliary evidence through orthology with a ciliary protein in another species. (iii) a non-human protein (sp2) can have a ciliary evidence through orthology with another species (sp3) and a link to OMIM through orthology with a human protein. In some cases, although linked to Sp2, the human and Sp3 proteins have no direct links, so that the human protein itself is not flagged as ciliary, whereas the associated OMIM entry is.
We wondered whether some of the 216 disorders in the filtered list could be ciliopathies and, after examination of the types of symptoms in the disease description and the kinds of ciliary evidences, 11 of these diseases can be proposed as candidate novel ciliopathies: four retinitis pigmentosa, a neuropathy, a neuroblastoma, a recessive deafness, a juvenile myoclonic epilepsy, an aldolase A deficiency, a sporadic breast cancer and a spinal muscular atrophy (see Table 1). Each disease can now be examined with the ciliary evidence in mind to check whether it could indeed be a ciliopathy.
Human genes associated with known and candidate ciliopathies
| Gene . | ID . | Band . | Synonyms . | MIM number . | Ciliary evidences . | Associated diseases . | Status . |
|---|---|---|---|---|---|---|---|
| AHI1 | ENSG00000135541 | 6q23.3 | JBTS3, AHI1, ORF1, FLJ20069 | 608894 | 1 | JBTS3 (608629), AHI1 (608629) | K |
| AIPL1 | ENSG00000129221 | 17p13.2 | AIPL1, LCA4 | 604392 | 3 | LCA4 (604393), AIPL1 (604393) | K |
| ALMS1 | ENSG00000116127 | 2p13.1 | ALMS1, KIAA0328 | 606844 | 2 | ALMS1 (203800) | K |
| ARL13B | ENSG00000169379 | 3q11.2 | ARL13B, ARL2L1, DKFZp761H079, JBTS8 | 608922 | 2 | JBTS8 (612291) | N |
| ARL6 | ENSG00000113966 | 3q11.2 | ARL6, BBS3 | 608845 | 6 | BBS3 (209900) | K |
| ARPKD | ENSG00000170927 | 6p12.2 | ARPKD, fibrocystin, tigmin, PKHD1, TIGM1, polyductin, FCYT | 606702 | 0 | PKHD1 (263200), ARPKD (263200) | K |
| BBS1 | ENSG00000174483 | 11q13.2 | BBS1, FLJ23590, DPP3 | 209901 | 6 | BBS1 (209900) | K |
| BBS10 | ENSG00000179941 | 12q21.2 | C12orf58, FLJ23560, BBS10 | 610148 | 0 | BBS10 (209900) | K |
| BBS11 | ENSG00000119401 | 9q33.1 | TRIM32, LGMD2H, HT2A, TATIP, BBS11 | 602290 | 0 | BBS11(209900) | K |
| BBS12 | ENSG00000181004 | 4q27 | FLJ35630, C4orf24, BBS12, FLJ41559 | 610683 | 0 | BBS12 (209900) | K |
| BBS2 | ENSG00000125124 | 16q12.2 | BBS2,BBS | 606151 | 5 | BBS2 (209900) | K |
| BBS4 | ENSG00000140463 | 15q24.1 | BBS4 | 600374 | 4 | BBS4 (209900) | K |
| BBS5 | ENSG00000163093 | 2q31.1 | DKFZp762I194, BBS5 | 603650 | 9 | BBS5 (209900) | K |
| BBS6 | ENSG00000125863 | 20p12.2 | MKKS, BBS6 | 604896 | 0 | BBS6 (209900) | K |
| BBS7 | ENSG00000138686 | 4q27 | BBS2L1, BBS7, FLJ10715 | 607590 | 5 | BBS7 (209900) | K |
| BBS9 | ENSG00000122507 | 7p14.3 | PTHB1, B1, BBS9 | 607968 | 5 | BBS9 (209900) | K |
| CCDC28B | ENSG00000160050 | 1p35.1 | CCDC28B, MGC1203, RP4-622L5.5 | 610162 | 0 | MGC1203 (209900) | N |
| CC2D2A | ENSG00000048342 | 4p15.33 | MKS6, NP_001073991.1, JBTS9, CC2D2A, KIAA1345 | 612013 | 6 | MKS6 (612284), JBTS9 (612285) | N |
| CEP290 | ENSG00000198707 | 12q21.32 | MKS4, JBTS5, SLSN6, BBS14, rd16, NPHP6, FLJ13615, KIAA0373, 3H11Ag, CEP290, LCA10 | 610142 | 3 | JBTS5 (610188), MKS4 (611134), SLSN6 (610189), BBS14 (209900), LCA10 (611755), NPHP6 | K |
| CRB1 | ENSG00000134376 | 1q31.3 | LCA8, RP12, CRB1 | 604210 | 0 | LCA8 (204000), RP12 (600105), CRB1 (604210) | K |
| CRX | ENSG00000105392 | 19q13.3 | CORD2, OTX3, CRD, LCA7, CRX | 602225 | 0 | CORD2 (120970), LCA7 (602225), RP (268000) | K |
| DNAH11 | ENSG00000105877 | 7p15.3 | DNHBL, DNAH11, DNAHBL, Dnahc11, CILD7, DPL11 | 603339 | 7 | CILD7 (611884) | K |
| DNAH5 | ENSG00000039139 | 5p15.2 | CILD3, PCD, KTGNR, HL1, Dnahc5, DNAH5 | 603335 | 8 | CILD3 (608644) | K |
| DNAI1 | ENSG00000122735 | 9p13.3 | DNAI1, PCD, CILD1 | 604366 | 9 | CILD1 (244400) | K |
| DNAI2 | ENSG00000171595 | 17q25.1 | DNAI2 | 605483 | 8 | CILD9 (612444) | N |
| GLIS2 | ENSG00000126603 | 16p13.3 | GLIS2, NPHP7 | 608539 | 0 | NPHP7 (611498) | K |
| GUCY2D | ENSG00000132518 | 17p13.1 | LCA1, GUCY2D, CYGD, CORD6, ROS-GC1, CORD5, GUC1A4, GUC2D, retGC, LCA, RETGC-1 | 600179 | 3 | LCA1 (204000), CORD6 (601777) | K |
| IFT80 | ENSG00000068885 | 3q25.33 | KIAA1374, WDR56, IFT80 | 611177 | 12 | ATD2 (611263) | K |
| IMPDH1 | ENSG00000106348 | 7q32.1 | sWSS2608, IMPDH1, RP10, LCA11 | 146690 | 1 | RP10 (180105), LCA11 (146690) | K |
| INVS | ENSG00000119509 | 9q31.1 | INVS, NPHP2 | 243305 | 0 | NPHP2 (602088) | K |
| IQCB1 | ENSG00000173226 | 3q13.33 | KIAA0036, IQCB1, NPHP5 | 609237 | 1 | NPHP5 (609254), SLN5 (609254) | K |
| KTU | ENSG00000165506 | 14q21.3 | KTU, C14orf104, FLJ10563 | 612517 | 0 | CILD10 (612518) | N |
| LCA5 | ENSG00000135338 | 6q14.1 | LCA5, C6orf152 | 611408 | 2 | LCA5 (604537) | K |
| MKS1 | ENSG00000011143 | 17q23.2 | FLJ20345, MKS, MKS1 | 609883 | 4 | MKS1 (249000), BBS13 (209900) | K |
| NEK8 | ENSG00000160602 | 17q11.2 | NEK8 | 609799 | 0 | NPHP9 | N |
| NPHP1 | ENSG00000144061 | 2q13 | NPHP1, NPH1, JBTS4 | 607100 | 4 | NPHP1 (256100), JBTS4 (609583), SLNS1 (266900) | K |
| NPHP3 | ENSG00000113971 | 3q22.1 | FLJ12592, NPH3, KIAA2000, NPHP3, FLJ30691, ACAD11, FLJ36696 | 608002 | 1 | NPHP3 (604387), SLNS3 (606995) | K |
| NPHP4 | ENSG00000131697 | 1p36.31 | NPHP4, nephroretinin, KIAA0673, SLSN4 | 607215 | 6 | NPHP4 (606996), SLSN4 (606996) | K |
| OFD1 | ENSG00000046651 | Xp22.2 | OFD1, CXorf5, 71-7A | 300170 | 1 | OFD1 (311200) | K |
| PKD1 | ENSG00000008710 | 16p13.3 | PBP, PKD1 | 601313 | 0 | PKD1 (173900) | K |
| PKD2 | ENSG00000118762 | 4q22.1 | PKD2, PKD4, PC2 | 173910 | 1 | PKD2 (173900) | K |
| RDH12 | ENSG00000139988 | 14q24.1 | FLJ30273, RDH12, SDR7C2 | 608830 | 1 | LCA13 (612712) | K |
| RPE65 | ENSG00000116745 | 1p31.2 | RP20, RPE65, rd12, LCA2 | 180069 | 1 | LCA2 (204100) | K |
| RPGRIP1 | ENSG00000092200 | 14q11.2 | RPGRIP1, LCA6, RGI1, RPGRIP | 605446 | 1 | CORD9 (608194), LCA6 (605446) | K |
| RPGRIP1L | ENSG00000103494 | 16q12.2 | MKS5, RPGRIP1L, JBTS7, NPHP8, CORS3, KIAA1005 | 610937 | 0 | MKS5 (611561), JBTS7 (611560), NPHP8, CORS3 | K |
| RSHL3, RSPH4a | ENSG00000111834 | 6q22.1 | RSHL3, dJ412I7.1, FLJ37974 | 612647 | 10 | CILD11 (612649) | N |
| RSPH9 | ENSG00000172426 | 6p21.1 | C6orf206, MRPS18AL1, FLJ30845 | 612648 | 8 | CILD12 (612650) | N |
| TMEM67 | ENSG00000164953 | 8q22.1 | MKS3, JBTS6, MGC26979, TMEM67 | 609884 | 1 | MKS3 (607361), JBTS6 (610688) | K |
| TTC8 | ENSG00000165533 | 14q32.11 | TTC8, BBS8 | 608132 | 7 | BBS8 (209900) | K |
| TXNDC3 | ENSG00000086288 | 7p14.1 | SPTRX2, CILD6, NME8, TXNDC3 | 607421 | 3 | CILD6 (610852) | K |
| ALDOA | ENSG00000149925 | 16p11.2 | ALDOA | 103850 | 3 | ALD1 (611881) | C |
| CCT5 | ENSG00000150753 | 5p15.2 | KIAA0098, CCT5 | 610150 | 5 | CCT5 (256840) | C |
| EFHC1 | ENSG00000096093 | 6p12.2 | EFHC1 | 608815 | 9 | JAE (607631) | C |
| NM23 | ENSG00000011052 | 17q21.33 | NM23-H2, NME1-NME2, NM23-H1, NME2, NM23 | 156490 | 5 | NB (256700) | C |
| PDE6A | ENSG00000132915 | 5q32 | PDEA, PDE6A | 180071 | 3 | RP43 (180071) | C |
| PDE6B | ENSG00000133256 | 4p16.3 | RP40, CSNB3, PDE6B, PDEB, rd1 | 180072 | 3 | RP40 (180072) | C |
| PHB1 | ENSG00000167085 | 17q21.33 | PHB, PHB1 | 176705 | 4 | PHB (176705) | C |
| PMCA2 | ENSG00000157087 | 3p25.3 | PMCA2, ATP2B2 | 108733 | 5 | ATP2B2 (601386) | C |
| RCNC2 | ENSG00000070729 | 16q13 | RCNC2, CNCG2, CNCG3L, GARP, CNGB1, GAR1, CNGB1B, RCNCb | 600724 | 2 | RP45 (268000) | C |
| TULP1 | ENSG00000112041 | 6p21.31 | RP14, TUBL1, TULP1 | 602280 | 4 | RP14 (600132) | C |
| UBE1X | ENSG00000130985 | Xp11.3 | UBE1X, A1S9T, UBA1, UBE1, GXP1 | 314370 | 4 | SMAX2 (301830) | C |
| Gene . | ID . | Band . | Synonyms . | MIM number . | Ciliary evidences . | Associated diseases . | Status . |
|---|---|---|---|---|---|---|---|
| AHI1 | ENSG00000135541 | 6q23.3 | JBTS3, AHI1, ORF1, FLJ20069 | 608894 | 1 | JBTS3 (608629), AHI1 (608629) | K |
| AIPL1 | ENSG00000129221 | 17p13.2 | AIPL1, LCA4 | 604392 | 3 | LCA4 (604393), AIPL1 (604393) | K |
| ALMS1 | ENSG00000116127 | 2p13.1 | ALMS1, KIAA0328 | 606844 | 2 | ALMS1 (203800) | K |
| ARL13B | ENSG00000169379 | 3q11.2 | ARL13B, ARL2L1, DKFZp761H079, JBTS8 | 608922 | 2 | JBTS8 (612291) | N |
| ARL6 | ENSG00000113966 | 3q11.2 | ARL6, BBS3 | 608845 | 6 | BBS3 (209900) | K |
| ARPKD | ENSG00000170927 | 6p12.2 | ARPKD, fibrocystin, tigmin, PKHD1, TIGM1, polyductin, FCYT | 606702 | 0 | PKHD1 (263200), ARPKD (263200) | K |
| BBS1 | ENSG00000174483 | 11q13.2 | BBS1, FLJ23590, DPP3 | 209901 | 6 | BBS1 (209900) | K |
| BBS10 | ENSG00000179941 | 12q21.2 | C12orf58, FLJ23560, BBS10 | 610148 | 0 | BBS10 (209900) | K |
| BBS11 | ENSG00000119401 | 9q33.1 | TRIM32, LGMD2H, HT2A, TATIP, BBS11 | 602290 | 0 | BBS11(209900) | K |
| BBS12 | ENSG00000181004 | 4q27 | FLJ35630, C4orf24, BBS12, FLJ41559 | 610683 | 0 | BBS12 (209900) | K |
| BBS2 | ENSG00000125124 | 16q12.2 | BBS2,BBS | 606151 | 5 | BBS2 (209900) | K |
| BBS4 | ENSG00000140463 | 15q24.1 | BBS4 | 600374 | 4 | BBS4 (209900) | K |
| BBS5 | ENSG00000163093 | 2q31.1 | DKFZp762I194, BBS5 | 603650 | 9 | BBS5 (209900) | K |
| BBS6 | ENSG00000125863 | 20p12.2 | MKKS, BBS6 | 604896 | 0 | BBS6 (209900) | K |
| BBS7 | ENSG00000138686 | 4q27 | BBS2L1, BBS7, FLJ10715 | 607590 | 5 | BBS7 (209900) | K |
| BBS9 | ENSG00000122507 | 7p14.3 | PTHB1, B1, BBS9 | 607968 | 5 | BBS9 (209900) | K |
| CCDC28B | ENSG00000160050 | 1p35.1 | CCDC28B, MGC1203, RP4-622L5.5 | 610162 | 0 | MGC1203 (209900) | N |
| CC2D2A | ENSG00000048342 | 4p15.33 | MKS6, NP_001073991.1, JBTS9, CC2D2A, KIAA1345 | 612013 | 6 | MKS6 (612284), JBTS9 (612285) | N |
| CEP290 | ENSG00000198707 | 12q21.32 | MKS4, JBTS5, SLSN6, BBS14, rd16, NPHP6, FLJ13615, KIAA0373, 3H11Ag, CEP290, LCA10 | 610142 | 3 | JBTS5 (610188), MKS4 (611134), SLSN6 (610189), BBS14 (209900), LCA10 (611755), NPHP6 | K |
| CRB1 | ENSG00000134376 | 1q31.3 | LCA8, RP12, CRB1 | 604210 | 0 | LCA8 (204000), RP12 (600105), CRB1 (604210) | K |
| CRX | ENSG00000105392 | 19q13.3 | CORD2, OTX3, CRD, LCA7, CRX | 602225 | 0 | CORD2 (120970), LCA7 (602225), RP (268000) | K |
| DNAH11 | ENSG00000105877 | 7p15.3 | DNHBL, DNAH11, DNAHBL, Dnahc11, CILD7, DPL11 | 603339 | 7 | CILD7 (611884) | K |
| DNAH5 | ENSG00000039139 | 5p15.2 | CILD3, PCD, KTGNR, HL1, Dnahc5, DNAH5 | 603335 | 8 | CILD3 (608644) | K |
| DNAI1 | ENSG00000122735 | 9p13.3 | DNAI1, PCD, CILD1 | 604366 | 9 | CILD1 (244400) | K |
| DNAI2 | ENSG00000171595 | 17q25.1 | DNAI2 | 605483 | 8 | CILD9 (612444) | N |
| GLIS2 | ENSG00000126603 | 16p13.3 | GLIS2, NPHP7 | 608539 | 0 | NPHP7 (611498) | K |
| GUCY2D | ENSG00000132518 | 17p13.1 | LCA1, GUCY2D, CYGD, CORD6, ROS-GC1, CORD5, GUC1A4, GUC2D, retGC, LCA, RETGC-1 | 600179 | 3 | LCA1 (204000), CORD6 (601777) | K |
| IFT80 | ENSG00000068885 | 3q25.33 | KIAA1374, WDR56, IFT80 | 611177 | 12 | ATD2 (611263) | K |
| IMPDH1 | ENSG00000106348 | 7q32.1 | sWSS2608, IMPDH1, RP10, LCA11 | 146690 | 1 | RP10 (180105), LCA11 (146690) | K |
| INVS | ENSG00000119509 | 9q31.1 | INVS, NPHP2 | 243305 | 0 | NPHP2 (602088) | K |
| IQCB1 | ENSG00000173226 | 3q13.33 | KIAA0036, IQCB1, NPHP5 | 609237 | 1 | NPHP5 (609254), SLN5 (609254) | K |
| KTU | ENSG00000165506 | 14q21.3 | KTU, C14orf104, FLJ10563 | 612517 | 0 | CILD10 (612518) | N |
| LCA5 | ENSG00000135338 | 6q14.1 | LCA5, C6orf152 | 611408 | 2 | LCA5 (604537) | K |
| MKS1 | ENSG00000011143 | 17q23.2 | FLJ20345, MKS, MKS1 | 609883 | 4 | MKS1 (249000), BBS13 (209900) | K |
| NEK8 | ENSG00000160602 | 17q11.2 | NEK8 | 609799 | 0 | NPHP9 | N |
| NPHP1 | ENSG00000144061 | 2q13 | NPHP1, NPH1, JBTS4 | 607100 | 4 | NPHP1 (256100), JBTS4 (609583), SLNS1 (266900) | K |
| NPHP3 | ENSG00000113971 | 3q22.1 | FLJ12592, NPH3, KIAA2000, NPHP3, FLJ30691, ACAD11, FLJ36696 | 608002 | 1 | NPHP3 (604387), SLNS3 (606995) | K |
| NPHP4 | ENSG00000131697 | 1p36.31 | NPHP4, nephroretinin, KIAA0673, SLSN4 | 607215 | 6 | NPHP4 (606996), SLSN4 (606996) | K |
| OFD1 | ENSG00000046651 | Xp22.2 | OFD1, CXorf5, 71-7A | 300170 | 1 | OFD1 (311200) | K |
| PKD1 | ENSG00000008710 | 16p13.3 | PBP, PKD1 | 601313 | 0 | PKD1 (173900) | K |
| PKD2 | ENSG00000118762 | 4q22.1 | PKD2, PKD4, PC2 | 173910 | 1 | PKD2 (173900) | K |
| RDH12 | ENSG00000139988 | 14q24.1 | FLJ30273, RDH12, SDR7C2 | 608830 | 1 | LCA13 (612712) | K |
| RPE65 | ENSG00000116745 | 1p31.2 | RP20, RPE65, rd12, LCA2 | 180069 | 1 | LCA2 (204100) | K |
| RPGRIP1 | ENSG00000092200 | 14q11.2 | RPGRIP1, LCA6, RGI1, RPGRIP | 605446 | 1 | CORD9 (608194), LCA6 (605446) | K |
| RPGRIP1L | ENSG00000103494 | 16q12.2 | MKS5, RPGRIP1L, JBTS7, NPHP8, CORS3, KIAA1005 | 610937 | 0 | MKS5 (611561), JBTS7 (611560), NPHP8, CORS3 | K |
| RSHL3, RSPH4a | ENSG00000111834 | 6q22.1 | RSHL3, dJ412I7.1, FLJ37974 | 612647 | 10 | CILD11 (612649) | N |
| RSPH9 | ENSG00000172426 | 6p21.1 | C6orf206, MRPS18AL1, FLJ30845 | 612648 | 8 | CILD12 (612650) | N |
| TMEM67 | ENSG00000164953 | 8q22.1 | MKS3, JBTS6, MGC26979, TMEM67 | 609884 | 1 | MKS3 (607361), JBTS6 (610688) | K |
| TTC8 | ENSG00000165533 | 14q32.11 | TTC8, BBS8 | 608132 | 7 | BBS8 (209900) | K |
| TXNDC3 | ENSG00000086288 | 7p14.1 | SPTRX2, CILD6, NME8, TXNDC3 | 607421 | 3 | CILD6 (610852) | K |
| ALDOA | ENSG00000149925 | 16p11.2 | ALDOA | 103850 | 3 | ALD1 (611881) | C |
| CCT5 | ENSG00000150753 | 5p15.2 | KIAA0098, CCT5 | 610150 | 5 | CCT5 (256840) | C |
| EFHC1 | ENSG00000096093 | 6p12.2 | EFHC1 | 608815 | 9 | JAE (607631) | C |
| NM23 | ENSG00000011052 | 17q21.33 | NM23-H2, NME1-NME2, NM23-H1, NME2, NM23 | 156490 | 5 | NB (256700) | C |
| PDE6A | ENSG00000132915 | 5q32 | PDEA, PDE6A | 180071 | 3 | RP43 (180071) | C |
| PDE6B | ENSG00000133256 | 4p16.3 | RP40, CSNB3, PDE6B, PDEB, rd1 | 180072 | 3 | RP40 (180072) | C |
| PHB1 | ENSG00000167085 | 17q21.33 | PHB, PHB1 | 176705 | 4 | PHB (176705) | C |
| PMCA2 | ENSG00000157087 | 3p25.3 | PMCA2, ATP2B2 | 108733 | 5 | ATP2B2 (601386) | C |
| RCNC2 | ENSG00000070729 | 16q13 | RCNC2, CNCG2, CNCG3L, GARP, CNGB1, GAR1, CNGB1B, RCNCb | 600724 | 2 | RP45 (268000) | C |
| TULP1 | ENSG00000112041 | 6p21.31 | RP14, TUBL1, TULP1 | 602280 | 4 | RP14 (600132) | C |
| UBE1X | ENSG00000130985 | Xp11.3 | UBE1X, A1S9T, UBA1, UBE1, GXP1 | 314370 | 4 | SMAX2 (301830) | C |
This table lists all the known or putative ciliopathies identified so far. In addition to the common gene name, the ID, the chromosomal location, the synonyms and the corresponding MIM number, we added the number of ciliary evidences (number of studies linked to the gene) and the diseases associated to deficiencies of the gene (names and MIM numbers). The last column refers to the status of the ciliopathy in our study; K: known ciliopathy referenced in OMIM; N: known ciliopathy not yet referenced in OMIM or not yet listed in the GeneMap section of OMIM, the tool that permits us to link diseases to genes; C: candidate novel ciliopathies.
Human genes associated with known and candidate ciliopathies
| Gene . | ID . | Band . | Synonyms . | MIM number . | Ciliary evidences . | Associated diseases . | Status . |
|---|---|---|---|---|---|---|---|
| AHI1 | ENSG00000135541 | 6q23.3 | JBTS3, AHI1, ORF1, FLJ20069 | 608894 | 1 | JBTS3 (608629), AHI1 (608629) | K |
| AIPL1 | ENSG00000129221 | 17p13.2 | AIPL1, LCA4 | 604392 | 3 | LCA4 (604393), AIPL1 (604393) | K |
| ALMS1 | ENSG00000116127 | 2p13.1 | ALMS1, KIAA0328 | 606844 | 2 | ALMS1 (203800) | K |
| ARL13B | ENSG00000169379 | 3q11.2 | ARL13B, ARL2L1, DKFZp761H079, JBTS8 | 608922 | 2 | JBTS8 (612291) | N |
| ARL6 | ENSG00000113966 | 3q11.2 | ARL6, BBS3 | 608845 | 6 | BBS3 (209900) | K |
| ARPKD | ENSG00000170927 | 6p12.2 | ARPKD, fibrocystin, tigmin, PKHD1, TIGM1, polyductin, FCYT | 606702 | 0 | PKHD1 (263200), ARPKD (263200) | K |
| BBS1 | ENSG00000174483 | 11q13.2 | BBS1, FLJ23590, DPP3 | 209901 | 6 | BBS1 (209900) | K |
| BBS10 | ENSG00000179941 | 12q21.2 | C12orf58, FLJ23560, BBS10 | 610148 | 0 | BBS10 (209900) | K |
| BBS11 | ENSG00000119401 | 9q33.1 | TRIM32, LGMD2H, HT2A, TATIP, BBS11 | 602290 | 0 | BBS11(209900) | K |
| BBS12 | ENSG00000181004 | 4q27 | FLJ35630, C4orf24, BBS12, FLJ41559 | 610683 | 0 | BBS12 (209900) | K |
| BBS2 | ENSG00000125124 | 16q12.2 | BBS2,BBS | 606151 | 5 | BBS2 (209900) | K |
| BBS4 | ENSG00000140463 | 15q24.1 | BBS4 | 600374 | 4 | BBS4 (209900) | K |
| BBS5 | ENSG00000163093 | 2q31.1 | DKFZp762I194, BBS5 | 603650 | 9 | BBS5 (209900) | K |
| BBS6 | ENSG00000125863 | 20p12.2 | MKKS, BBS6 | 604896 | 0 | BBS6 (209900) | K |
| BBS7 | ENSG00000138686 | 4q27 | BBS2L1, BBS7, FLJ10715 | 607590 | 5 | BBS7 (209900) | K |
| BBS9 | ENSG00000122507 | 7p14.3 | PTHB1, B1, BBS9 | 607968 | 5 | BBS9 (209900) | K |
| CCDC28B | ENSG00000160050 | 1p35.1 | CCDC28B, MGC1203, RP4-622L5.5 | 610162 | 0 | MGC1203 (209900) | N |
| CC2D2A | ENSG00000048342 | 4p15.33 | MKS6, NP_001073991.1, JBTS9, CC2D2A, KIAA1345 | 612013 | 6 | MKS6 (612284), JBTS9 (612285) | N |
| CEP290 | ENSG00000198707 | 12q21.32 | MKS4, JBTS5, SLSN6, BBS14, rd16, NPHP6, FLJ13615, KIAA0373, 3H11Ag, CEP290, LCA10 | 610142 | 3 | JBTS5 (610188), MKS4 (611134), SLSN6 (610189), BBS14 (209900), LCA10 (611755), NPHP6 | K |
| CRB1 | ENSG00000134376 | 1q31.3 | LCA8, RP12, CRB1 | 604210 | 0 | LCA8 (204000), RP12 (600105), CRB1 (604210) | K |
| CRX | ENSG00000105392 | 19q13.3 | CORD2, OTX3, CRD, LCA7, CRX | 602225 | 0 | CORD2 (120970), LCA7 (602225), RP (268000) | K |
| DNAH11 | ENSG00000105877 | 7p15.3 | DNHBL, DNAH11, DNAHBL, Dnahc11, CILD7, DPL11 | 603339 | 7 | CILD7 (611884) | K |
| DNAH5 | ENSG00000039139 | 5p15.2 | CILD3, PCD, KTGNR, HL1, Dnahc5, DNAH5 | 603335 | 8 | CILD3 (608644) | K |
| DNAI1 | ENSG00000122735 | 9p13.3 | DNAI1, PCD, CILD1 | 604366 | 9 | CILD1 (244400) | K |
| DNAI2 | ENSG00000171595 | 17q25.1 | DNAI2 | 605483 | 8 | CILD9 (612444) | N |
| GLIS2 | ENSG00000126603 | 16p13.3 | GLIS2, NPHP7 | 608539 | 0 | NPHP7 (611498) | K |
| GUCY2D | ENSG00000132518 | 17p13.1 | LCA1, GUCY2D, CYGD, CORD6, ROS-GC1, CORD5, GUC1A4, GUC2D, retGC, LCA, RETGC-1 | 600179 | 3 | LCA1 (204000), CORD6 (601777) | K |
| IFT80 | ENSG00000068885 | 3q25.33 | KIAA1374, WDR56, IFT80 | 611177 | 12 | ATD2 (611263) | K |
| IMPDH1 | ENSG00000106348 | 7q32.1 | sWSS2608, IMPDH1, RP10, LCA11 | 146690 | 1 | RP10 (180105), LCA11 (146690) | K |
| INVS | ENSG00000119509 | 9q31.1 | INVS, NPHP2 | 243305 | 0 | NPHP2 (602088) | K |
| IQCB1 | ENSG00000173226 | 3q13.33 | KIAA0036, IQCB1, NPHP5 | 609237 | 1 | NPHP5 (609254), SLN5 (609254) | K |
| KTU | ENSG00000165506 | 14q21.3 | KTU, C14orf104, FLJ10563 | 612517 | 0 | CILD10 (612518) | N |
| LCA5 | ENSG00000135338 | 6q14.1 | LCA5, C6orf152 | 611408 | 2 | LCA5 (604537) | K |
| MKS1 | ENSG00000011143 | 17q23.2 | FLJ20345, MKS, MKS1 | 609883 | 4 | MKS1 (249000), BBS13 (209900) | K |
| NEK8 | ENSG00000160602 | 17q11.2 | NEK8 | 609799 | 0 | NPHP9 | N |
| NPHP1 | ENSG00000144061 | 2q13 | NPHP1, NPH1, JBTS4 | 607100 | 4 | NPHP1 (256100), JBTS4 (609583), SLNS1 (266900) | K |
| NPHP3 | ENSG00000113971 | 3q22.1 | FLJ12592, NPH3, KIAA2000, NPHP3, FLJ30691, ACAD11, FLJ36696 | 608002 | 1 | NPHP3 (604387), SLNS3 (606995) | K |
| NPHP4 | ENSG00000131697 | 1p36.31 | NPHP4, nephroretinin, KIAA0673, SLSN4 | 607215 | 6 | NPHP4 (606996), SLSN4 (606996) | K |
| OFD1 | ENSG00000046651 | Xp22.2 | OFD1, CXorf5, 71-7A | 300170 | 1 | OFD1 (311200) | K |
| PKD1 | ENSG00000008710 | 16p13.3 | PBP, PKD1 | 601313 | 0 | PKD1 (173900) | K |
| PKD2 | ENSG00000118762 | 4q22.1 | PKD2, PKD4, PC2 | 173910 | 1 | PKD2 (173900) | K |
| RDH12 | ENSG00000139988 | 14q24.1 | FLJ30273, RDH12, SDR7C2 | 608830 | 1 | LCA13 (612712) | K |
| RPE65 | ENSG00000116745 | 1p31.2 | RP20, RPE65, rd12, LCA2 | 180069 | 1 | LCA2 (204100) | K |
| RPGRIP1 | ENSG00000092200 | 14q11.2 | RPGRIP1, LCA6, RGI1, RPGRIP | 605446 | 1 | CORD9 (608194), LCA6 (605446) | K |
| RPGRIP1L | ENSG00000103494 | 16q12.2 | MKS5, RPGRIP1L, JBTS7, NPHP8, CORS3, KIAA1005 | 610937 | 0 | MKS5 (611561), JBTS7 (611560), NPHP8, CORS3 | K |
| RSHL3, RSPH4a | ENSG00000111834 | 6q22.1 | RSHL3, dJ412I7.1, FLJ37974 | 612647 | 10 | CILD11 (612649) | N |
| RSPH9 | ENSG00000172426 | 6p21.1 | C6orf206, MRPS18AL1, FLJ30845 | 612648 | 8 | CILD12 (612650) | N |
| TMEM67 | ENSG00000164953 | 8q22.1 | MKS3, JBTS6, MGC26979, TMEM67 | 609884 | 1 | MKS3 (607361), JBTS6 (610688) | K |
| TTC8 | ENSG00000165533 | 14q32.11 | TTC8, BBS8 | 608132 | 7 | BBS8 (209900) | K |
| TXNDC3 | ENSG00000086288 | 7p14.1 | SPTRX2, CILD6, NME8, TXNDC3 | 607421 | 3 | CILD6 (610852) | K |
| ALDOA | ENSG00000149925 | 16p11.2 | ALDOA | 103850 | 3 | ALD1 (611881) | C |
| CCT5 | ENSG00000150753 | 5p15.2 | KIAA0098, CCT5 | 610150 | 5 | CCT5 (256840) | C |
| EFHC1 | ENSG00000096093 | 6p12.2 | EFHC1 | 608815 | 9 | JAE (607631) | C |
| NM23 | ENSG00000011052 | 17q21.33 | NM23-H2, NME1-NME2, NM23-H1, NME2, NM23 | 156490 | 5 | NB (256700) | C |
| PDE6A | ENSG00000132915 | 5q32 | PDEA, PDE6A | 180071 | 3 | RP43 (180071) | C |
| PDE6B | ENSG00000133256 | 4p16.3 | RP40, CSNB3, PDE6B, PDEB, rd1 | 180072 | 3 | RP40 (180072) | C |
| PHB1 | ENSG00000167085 | 17q21.33 | PHB, PHB1 | 176705 | 4 | PHB (176705) | C |
| PMCA2 | ENSG00000157087 | 3p25.3 | PMCA2, ATP2B2 | 108733 | 5 | ATP2B2 (601386) | C |
| RCNC2 | ENSG00000070729 | 16q13 | RCNC2, CNCG2, CNCG3L, GARP, CNGB1, GAR1, CNGB1B, RCNCb | 600724 | 2 | RP45 (268000) | C |
| TULP1 | ENSG00000112041 | 6p21.31 | RP14, TUBL1, TULP1 | 602280 | 4 | RP14 (600132) | C |
| UBE1X | ENSG00000130985 | Xp11.3 | UBE1X, A1S9T, UBA1, UBE1, GXP1 | 314370 | 4 | SMAX2 (301830) | C |
| Gene . | ID . | Band . | Synonyms . | MIM number . | Ciliary evidences . | Associated diseases . | Status . |
|---|---|---|---|---|---|---|---|
| AHI1 | ENSG00000135541 | 6q23.3 | JBTS3, AHI1, ORF1, FLJ20069 | 608894 | 1 | JBTS3 (608629), AHI1 (608629) | K |
| AIPL1 | ENSG00000129221 | 17p13.2 | AIPL1, LCA4 | 604392 | 3 | LCA4 (604393), AIPL1 (604393) | K |
| ALMS1 | ENSG00000116127 | 2p13.1 | ALMS1, KIAA0328 | 606844 | 2 | ALMS1 (203800) | K |
| ARL13B | ENSG00000169379 | 3q11.2 | ARL13B, ARL2L1, DKFZp761H079, JBTS8 | 608922 | 2 | JBTS8 (612291) | N |
| ARL6 | ENSG00000113966 | 3q11.2 | ARL6, BBS3 | 608845 | 6 | BBS3 (209900) | K |
| ARPKD | ENSG00000170927 | 6p12.2 | ARPKD, fibrocystin, tigmin, PKHD1, TIGM1, polyductin, FCYT | 606702 | 0 | PKHD1 (263200), ARPKD (263200) | K |
| BBS1 | ENSG00000174483 | 11q13.2 | BBS1, FLJ23590, DPP3 | 209901 | 6 | BBS1 (209900) | K |
| BBS10 | ENSG00000179941 | 12q21.2 | C12orf58, FLJ23560, BBS10 | 610148 | 0 | BBS10 (209900) | K |
| BBS11 | ENSG00000119401 | 9q33.1 | TRIM32, LGMD2H, HT2A, TATIP, BBS11 | 602290 | 0 | BBS11(209900) | K |
| BBS12 | ENSG00000181004 | 4q27 | FLJ35630, C4orf24, BBS12, FLJ41559 | 610683 | 0 | BBS12 (209900) | K |
| BBS2 | ENSG00000125124 | 16q12.2 | BBS2,BBS | 606151 | 5 | BBS2 (209900) | K |
| BBS4 | ENSG00000140463 | 15q24.1 | BBS4 | 600374 | 4 | BBS4 (209900) | K |
| BBS5 | ENSG00000163093 | 2q31.1 | DKFZp762I194, BBS5 | 603650 | 9 | BBS5 (209900) | K |
| BBS6 | ENSG00000125863 | 20p12.2 | MKKS, BBS6 | 604896 | 0 | BBS6 (209900) | K |
| BBS7 | ENSG00000138686 | 4q27 | BBS2L1, BBS7, FLJ10715 | 607590 | 5 | BBS7 (209900) | K |
| BBS9 | ENSG00000122507 | 7p14.3 | PTHB1, B1, BBS9 | 607968 | 5 | BBS9 (209900) | K |
| CCDC28B | ENSG00000160050 | 1p35.1 | CCDC28B, MGC1203, RP4-622L5.5 | 610162 | 0 | MGC1203 (209900) | N |
| CC2D2A | ENSG00000048342 | 4p15.33 | MKS6, NP_001073991.1, JBTS9, CC2D2A, KIAA1345 | 612013 | 6 | MKS6 (612284), JBTS9 (612285) | N |
| CEP290 | ENSG00000198707 | 12q21.32 | MKS4, JBTS5, SLSN6, BBS14, rd16, NPHP6, FLJ13615, KIAA0373, 3H11Ag, CEP290, LCA10 | 610142 | 3 | JBTS5 (610188), MKS4 (611134), SLSN6 (610189), BBS14 (209900), LCA10 (611755), NPHP6 | K |
| CRB1 | ENSG00000134376 | 1q31.3 | LCA8, RP12, CRB1 | 604210 | 0 | LCA8 (204000), RP12 (600105), CRB1 (604210) | K |
| CRX | ENSG00000105392 | 19q13.3 | CORD2, OTX3, CRD, LCA7, CRX | 602225 | 0 | CORD2 (120970), LCA7 (602225), RP (268000) | K |
| DNAH11 | ENSG00000105877 | 7p15.3 | DNHBL, DNAH11, DNAHBL, Dnahc11, CILD7, DPL11 | 603339 | 7 | CILD7 (611884) | K |
| DNAH5 | ENSG00000039139 | 5p15.2 | CILD3, PCD, KTGNR, HL1, Dnahc5, DNAH5 | 603335 | 8 | CILD3 (608644) | K |
| DNAI1 | ENSG00000122735 | 9p13.3 | DNAI1, PCD, CILD1 | 604366 | 9 | CILD1 (244400) | K |
| DNAI2 | ENSG00000171595 | 17q25.1 | DNAI2 | 605483 | 8 | CILD9 (612444) | N |
| GLIS2 | ENSG00000126603 | 16p13.3 | GLIS2, NPHP7 | 608539 | 0 | NPHP7 (611498) | K |
| GUCY2D | ENSG00000132518 | 17p13.1 | LCA1, GUCY2D, CYGD, CORD6, ROS-GC1, CORD5, GUC1A4, GUC2D, retGC, LCA, RETGC-1 | 600179 | 3 | LCA1 (204000), CORD6 (601777) | K |
| IFT80 | ENSG00000068885 | 3q25.33 | KIAA1374, WDR56, IFT80 | 611177 | 12 | ATD2 (611263) | K |
| IMPDH1 | ENSG00000106348 | 7q32.1 | sWSS2608, IMPDH1, RP10, LCA11 | 146690 | 1 | RP10 (180105), LCA11 (146690) | K |
| INVS | ENSG00000119509 | 9q31.1 | INVS, NPHP2 | 243305 | 0 | NPHP2 (602088) | K |
| IQCB1 | ENSG00000173226 | 3q13.33 | KIAA0036, IQCB1, NPHP5 | 609237 | 1 | NPHP5 (609254), SLN5 (609254) | K |
| KTU | ENSG00000165506 | 14q21.3 | KTU, C14orf104, FLJ10563 | 612517 | 0 | CILD10 (612518) | N |
| LCA5 | ENSG00000135338 | 6q14.1 | LCA5, C6orf152 | 611408 | 2 | LCA5 (604537) | K |
| MKS1 | ENSG00000011143 | 17q23.2 | FLJ20345, MKS, MKS1 | 609883 | 4 | MKS1 (249000), BBS13 (209900) | K |
| NEK8 | ENSG00000160602 | 17q11.2 | NEK8 | 609799 | 0 | NPHP9 | N |
| NPHP1 | ENSG00000144061 | 2q13 | NPHP1, NPH1, JBTS4 | 607100 | 4 | NPHP1 (256100), JBTS4 (609583), SLNS1 (266900) | K |
| NPHP3 | ENSG00000113971 | 3q22.1 | FLJ12592, NPH3, KIAA2000, NPHP3, FLJ30691, ACAD11, FLJ36696 | 608002 | 1 | NPHP3 (604387), SLNS3 (606995) | K |
| NPHP4 | ENSG00000131697 | 1p36.31 | NPHP4, nephroretinin, KIAA0673, SLSN4 | 607215 | 6 | NPHP4 (606996), SLSN4 (606996) | K |
| OFD1 | ENSG00000046651 | Xp22.2 | OFD1, CXorf5, 71-7A | 300170 | 1 | OFD1 (311200) | K |
| PKD1 | ENSG00000008710 | 16p13.3 | PBP, PKD1 | 601313 | 0 | PKD1 (173900) | K |
| PKD2 | ENSG00000118762 | 4q22.1 | PKD2, PKD4, PC2 | 173910 | 1 | PKD2 (173900) | K |
| RDH12 | ENSG00000139988 | 14q24.1 | FLJ30273, RDH12, SDR7C2 | 608830 | 1 | LCA13 (612712) | K |
| RPE65 | ENSG00000116745 | 1p31.2 | RP20, RPE65, rd12, LCA2 | 180069 | 1 | LCA2 (204100) | K |
| RPGRIP1 | ENSG00000092200 | 14q11.2 | RPGRIP1, LCA6, RGI1, RPGRIP | 605446 | 1 | CORD9 (608194), LCA6 (605446) | K |
| RPGRIP1L | ENSG00000103494 | 16q12.2 | MKS5, RPGRIP1L, JBTS7, NPHP8, CORS3, KIAA1005 | 610937 | 0 | MKS5 (611561), JBTS7 (611560), NPHP8, CORS3 | K |
| RSHL3, RSPH4a | ENSG00000111834 | 6q22.1 | RSHL3, dJ412I7.1, FLJ37974 | 612647 | 10 | CILD11 (612649) | N |
| RSPH9 | ENSG00000172426 | 6p21.1 | C6orf206, MRPS18AL1, FLJ30845 | 612648 | 8 | CILD12 (612650) | N |
| TMEM67 | ENSG00000164953 | 8q22.1 | MKS3, JBTS6, MGC26979, TMEM67 | 609884 | 1 | MKS3 (607361), JBTS6 (610688) | K |
| TTC8 | ENSG00000165533 | 14q32.11 | TTC8, BBS8 | 608132 | 7 | BBS8 (209900) | K |
| TXNDC3 | ENSG00000086288 | 7p14.1 | SPTRX2, CILD6, NME8, TXNDC3 | 607421 | 3 | CILD6 (610852) | K |
| ALDOA | ENSG00000149925 | 16p11.2 | ALDOA | 103850 | 3 | ALD1 (611881) | C |
| CCT5 | ENSG00000150753 | 5p15.2 | KIAA0098, CCT5 | 610150 | 5 | CCT5 (256840) | C |
| EFHC1 | ENSG00000096093 | 6p12.2 | EFHC1 | 608815 | 9 | JAE (607631) | C |
| NM23 | ENSG00000011052 | 17q21.33 | NM23-H2, NME1-NME2, NM23-H1, NME2, NM23 | 156490 | 5 | NB (256700) | C |
| PDE6A | ENSG00000132915 | 5q32 | PDEA, PDE6A | 180071 | 3 | RP43 (180071) | C |
| PDE6B | ENSG00000133256 | 4p16.3 | RP40, CSNB3, PDE6B, PDEB, rd1 | 180072 | 3 | RP40 (180072) | C |
| PHB1 | ENSG00000167085 | 17q21.33 | PHB, PHB1 | 176705 | 4 | PHB (176705) | C |
| PMCA2 | ENSG00000157087 | 3p25.3 | PMCA2, ATP2B2 | 108733 | 5 | ATP2B2 (601386) | C |
| RCNC2 | ENSG00000070729 | 16q13 | RCNC2, CNCG2, CNCG3L, GARP, CNGB1, GAR1, CNGB1B, RCNCb | 600724 | 2 | RP45 (268000) | C |
| TULP1 | ENSG00000112041 | 6p21.31 | RP14, TUBL1, TULP1 | 602280 | 4 | RP14 (600132) | C |
| UBE1X | ENSG00000130985 | Xp11.3 | UBE1X, A1S9T, UBA1, UBE1, GXP1 | 314370 | 4 | SMAX2 (301830) | C |
This table lists all the known or putative ciliopathies identified so far. In addition to the common gene name, the ID, the chromosomal location, the synonyms and the corresponding MIM number, we added the number of ciliary evidences (number of studies linked to the gene) and the diseases associated to deficiencies of the gene (names and MIM numbers). The last column refers to the status of the ciliopathy in our study; K: known ciliopathy referenced in OMIM; N: known ciliopathy not yet referenced in OMIM or not yet listed in the GeneMap section of OMIM, the tool that permits us to link diseases to genes; C: candidate novel ciliopathies.
Finally, several hundred orphan diseases exist in man, with symptoms that may evoke a ciliary origin (deafness, retinal defects, obesity, polydactyly, kidney polycystosis, mental retardation), but with genetic location on chromosomes only imprecisely determined by linkage with markers. In Cildb, we have built a special section, the ‘Hsapiens OMIM database’, in which human proteins and OMIM entries, even imprecisely localized, can be displayed. For instance, the ciliopathy ‘Senior-Loken syndrome 3 (SLSN3)’ (OMIM 606995) is localized to chromosome 3, region q22, thus encompassing 831 genes. These genes with their attributes can be displayed to help find candidates for this syndrome.
Conclusion—future challenges
The generation of new high throughput ciliary data from Paramecium prompted us to build Cildb, a new database that integrates heterogeneous information from a variety of sources. The versatility of Cildb makes it a valuable knowledgebase, allowing queries from any proteome and using raw data from high throughput ciliary studies and multi-criteria queries. Cildb also permits the identification of ciliopathy genes and can even help identify candidate genes for diseases imprecisely localized on chromosomes. Beyond ciliary data, Cildb contains easy to retrieve information pertinent for general analyses of proteins, comparative genomics and linking to OMIM data concerning human genetic disorders.
Although updating Cildb is computer-time consuming, the procedure is straightforward. The next challenge will be to incorporate additional information necessary for ontology-aware phenotype descriptions (33). This would allow us to add high throughput RNAi studies or genetic screens, such as the ones conducted in the fish Danio reirio (34).
Funding
CNRS and the Agence Nationale de la Recherche, grant number NT05-2_41522. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Acknowledgements
The authors are grateful to Anne Laurençon who spent time testing Cildb and participated in its improvement and to the INRA MIGALE bioinformatics platform for providing computational resources.



![Orthology calculations and links to ciliary studies underlying Cildb. The 18 species analyzed are represented as circle arcs proportional to the size of the proteome, in red when ciliary studies exist (delineated as ticks outside the circle), in orange when centriole/cilia exist, but without high throughput studies, and in blue when no cilia or centriole exists (At: Arabidopsis thaliana; Ce: Caenorhabditis elegans; Ci: Ciona intestinalis; Cr: Chlamydomonas reinhardtii; Dd: Dictyostelium discoideum; Dm: Drosophila melanogaster; Dr: Danio rerio; Ec: Escherichia coli; Gl: Giardia lamblia; Hs: Homo sapiens; Mm: Mus musculus; Pf: Plasmodium falciparum; Pt: Paramecium tetraurelia; Rn: Rattus norvegicus; Sc: Saccharomyces cerevisiae; Sp: Schizosaccharomyces pombe; Tb: Trypanosoma brucei; Tt: Tetrahymena thermophila). The colors of the ticks for the studies are explained in the inset to distinguish proteomic, transcriptomic and comparative genomic studies. Andersen (35); Arnaiz [this article for proteomics, or Arnaiz et al. (in preparation) for transcriptome analysis]; Avidor-Reiss (36); Blacque (37); Broadhead (38); Chen (39); Efimenko (40); Keller (41); Kilburn (42); Laurençon (43); Li (16); Liu (44); Mayer (45); Ostrowski (19); Pazour (26); Smith (18); Stolc (46). For each species, the whole proteome was compared by BLASTp to itself and to all the other ones, in order to apply our three orthology filters (Inparanoid, Inparanoid + filtered best hits, BLASTp cutoff), as described in experimental procedures. The high throughput ciliary studies were then remapped to the proteins, so that links between orthology and ciliary properties are created. The genome versions and the origin of the ciliary study data in Cildb are reported in Supplementary Table S1. The human genome is also linked to the OMIM database in which genetic disorders appear.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/database/2009/10.1093_database_bap022/3/m_bap022f1.jpeg?Expires=1716312671&Signature=lM~uanOdQEmVtSjslJKfCL6ZIF8FQoFFoKXv2BF57bWPpn-nvgufsVPXGb072Ijafa8rCinIEanshovqc7uXlVGV25gmPRHBljVqiv4CCbRbW2VQ3V6ka0EypByFzdnrQPWzj00B~VIbUQ9cwyPhxErg-9Knn-T7M65qmd5Vza06V-idRTokg0l9olt4q5TqRxheD-9POxb356c3-RAkfVvd7OUqUZGgwYUrfLQ7fHVgIRYv-DvFHvCT2uZTg6P3je7YxfJ9EKQlTaWpOl~uHtLpNsUosRZVhHptJ9JvAYvw6b3bu2bvIvlrbev60xKLVNyxoymdyj31VVc0Rwu48w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Comparison of ciliary proteomic studies from different unicellular models. Using Cildb, we compared the proteomic studies of purified cilia/flagella of Chalmydomonas, Paramecium, Tetrahymena and Trypanosoma. The protocol was the same for each study: (i) Build a BioMart ‘new query’ in Cildb using the ‘Inparanoid and filtered best hits’ as homology method. (ii) Choose the species in which the ciliary study examined was conducted (e.g. Chlamydomonas). (iii) Filter proteins identified in the relevant proteomic study (e.g. Pazour’s proteomics) with medium stringency confidence (two or more different peptides). (iv) Select the ‘number of ciliary studies in all species with medium confidence’ as an attribute. (v) Count and display the results as xls tables. In the first column of each graph, the total height represents the number of proteins identified in the study, the dark blue proteins are those found as ciliary in at least another study, pale blue proteins are those found as ciliary only in this study, and grey proteins are those annotated as ribosomal proteins or histones, representing likely contaminants of the ciliary preparation. (vi) Filter proteins using the same criteria as in (iii) as well as the existence of ‘orthologs’ in each species used in this comparison (the other protists plus human). This represents as many queries as species examined and gives the height of the green bars for each species (the grey part corresponding to ribosomal proteins and histones). (vii) Filter as in (vi) and look in the results for proteins identified by ciliary proteomics in the target species [e.g. this study for Paramecium or Broadhead et al. (38) for Trypanosoma], represented as dark green, whether they have been found as ciliary in other species (medium green) or not (pale green).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/database/2009/10.1093_database_bap022/3/m_bap022f4.jpeg?Expires=1716312671&Signature=D7pWz295eAqAVww9a-lb2k~NhRlJVllZPqmLNyagDCz1cyxN3C-P9SDco9R0WKeZEv3YXYquAlSlIl9bJCOndK06b5Yqw73hklaA6bxLIIg4PNhXAqsuSOZAGYxXcyhq7HQsIOYgyu6Nu2RJ7KNzS6Gu3xt4Bjm4kL~SQgpi1s5LBiR0TUhHfw6RLVm61vw2fL0EN9L1WZUJYk344AjGznnegcSqWTrmdu7cFwjDypCAi6zDp7oGEm3PJB1gYUlXdxU55tLD-ENDagyU2e72X5e8g0Y8NeWroG5OJBNPtXuQq3TKiRJ20Sow2C6lrjkEx9clJ1gFZq8ZID2XarAWdA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)