-
PDF
- Split View
-
Views
-
Cite
Cite
François Moreews, Gaelle Rauffet, Patrice Dehais, Christophe Klopp, SigReannot-mart: a query environment for expression microarray probe re-annotations, Database, Volume 2011, 2011, bar025, https://doi.org/10.1093/database/bar025
Close - Share Icon Share
Abstract
Expression microarrays are commonly used to study transcriptomes. Most of the arrays are now based on oligo-nucleotide probes. Probe design being a tedious task, it often takes place once at the beginning of the project. The oligo set is then used for several years. During this time period, the knowledge gathered by the community on the genome and the transcriptome increases and gets more precise. Therefore re-annotating the set is essential to supply the biologists with up-to-date annotations. SigReannot-mart is a query environment populated with regularly updated annotations for different oligo sets. It stores the results of the SigReannot pipeline that has mainly been used on farm and aquaculture species. It permits easy extraction in different formats using filters. It is used to compare probe sets on different criteria, to choose the set for a given experiment to mix probe sets in order to create a new one.
Database URL:http://sigreannot-mart.toulouse.inra.fr/
Project description
Updating annotations
The main step of the microarray creation process is the probe design. The design tools aim at maximizing the probe sequence specificity while conserving the largest possible covered transcript or gene set. The genome and transcriptome sequences used as references during the design evolve with each new assembly or new gene build, modifying the link between the probes and the corresponding biological entities. As nearly, all the annotations related to a probe are based on this link, the annotation interpretation could become hazardous just a few months after the design, especially for organisms with an unfinished genome or a partially known transcriptome, like many farm animal and aquaculture species.
Consequently, researchers will need updated annotations based on the best possible probe-transcript link. This is the first goal of the SigReannot-mart database.
Re-assessing probe specificity
A specificity indicator is produced by the pipeline (1) and stored for each probe, this criteria can be used to evaluate the specificity of a probe and its chance of multiple transcripts crosshybridization. It is based on the number and the similarity scores of the blastn (2) alignments between a probe and a transcript or a genomic location.
Like in OligoRAP (3) and IMAD (4) pipelines, probes are assigned to different Target Specificity Classes (Table 1), based on the amount and type of hits (5).
Probe target specificity classes (TSCs) and subclasses
| TSCs . | Description . |
|---|---|
| 1 | One good hit and no noise |
| 2 | One good hit with noise |
| 3 | No hit, one noise ≥30 bp |
| 4 | No hit, one noise ≥20 and <30 bp |
| 5 | No hit many noises |
| 6 | No good hit no noises |
| 7 | Many good hits |
| 7.1 (subclasses) | Many good hits but one entity |
| MH (subclasses) | Multiple hits on one chromosome |
| MC (subclasses) | Hits on multiple chromosomes |
| TSCs . | Description . |
|---|---|
| 1 | One good hit and no noise |
| 2 | One good hit with noise |
| 3 | No hit, one noise ≥30 bp |
| 4 | No hit, one noise ≥20 and <30 bp |
| 5 | No hit many noises |
| 6 | No good hit no noises |
| 7 | Many good hits |
| 7.1 (subclasses) | Many good hits but one entity |
| MH (subclasses) | Multiple hits on one chromosome |
| MC (subclasses) | Hits on multiple chromosomes |
These quality indicators use a Blastn similarity search on the transcriptome of the studied species. As an illustration, the users can decide to reject the probes with many hits (Category 7) or not, using a complementary fine grain subcategory indicator (7.1, MH, MC). Ensembl provides probe set mapping but do not provides TSCs and stores only probes with one genomic hit and no more than one mismatch.
Probe target specificity classes (TSCs) and subclasses
| TSCs . | Description . |
|---|---|
| 1 | One good hit and no noise |
| 2 | One good hit with noise |
| 3 | No hit, one noise ≥30 bp |
| 4 | No hit, one noise ≥20 and <30 bp |
| 5 | No hit many noises |
| 6 | No good hit no noises |
| 7 | Many good hits |
| 7.1 (subclasses) | Many good hits but one entity |
| MH (subclasses) | Multiple hits on one chromosome |
| MC (subclasses) | Hits on multiple chromosomes |
| TSCs . | Description . |
|---|---|
| 1 | One good hit and no noise |
| 2 | One good hit with noise |
| 3 | No hit, one noise ≥30 bp |
| 4 | No hit, one noise ≥20 and <30 bp |
| 5 | No hit many noises |
| 6 | No good hit no noises |
| 7 | Many good hits |
| 7.1 (subclasses) | Many good hits but one entity |
| MH (subclasses) | Multiple hits on one chromosome |
| MC (subclasses) | Hits on multiple chromosomes |
These quality indicators use a Blastn similarity search on the transcriptome of the studied species. As an illustration, the users can decide to reject the probes with many hits (Category 7) or not, using a complementary fine grain subcategory indicator (7.1, MH, MC). Ensembl provides probe set mapping but do not provides TSCs and stores only probes with one genomic hit and no more than one mismatch.
The user can decide the level of probe specificity he wants to focus his analysis on. The specificity indicator is of high interest for biologists when they start interpreting their list of over- and under expressed genes. For the annotation of unspecific probes, only a more thorough analysis will help to understand which of the biological entities is the source of the signal. This is why SigReannot-mart provides complementary specificity subcategories that use hit location and description (Table 1).
Providing rich annotation using multiple data sources
Usually microarray manufacturers provide annotation files for their oligo sets. But they do not up-date these files with each new version of the genome annotation produced by Ensembl or the NCBI. These annotation files often contain limited information such as gene name, genomic location and some Gene Ontology (GO) terms. To help scientists interpret microarray expression data, sigReannot-mart integrates multiple complementary data sources covering external references, orthologs genes and pathways. External reference and orthologous genes are of high interest for the extraction of pathway information from sources such as David (6) or IPA (7).
Comparing array designs
When a biologist needs to choose between different available array designs, a common comparison criterion is the transcriptome coverage of the probe set (8,9). It can be evaluated by extracting the lists of pathways, GO terms, transcripts and gene IDs related to each set. But such a task tends to be tedious when the oligo sets are not annotated using the same methods and data sources. By giving access to shared standardized microarray annotations, sigReannot-mart addresses this need.
Designing new oligo sets from existing ones
Thanks to new printing techniques, it has been quite common for a number of years to build custom gene expression microarrays, even for a single scientist or a research team. The design step being a harsh time-consuming task, the strategy to build these custom microarrays is often to select probes from different existing platforms. This strategy also permits to verify the expression range of each oligo-nucleotide in the available data sets. SigReannot-mart can be used to simplify this task: the probes coming from different sets, sharing the same annotation process, can easily be selected, merged and compared to all the predicted transcripts of a studied species in order to generate the new set, with better transcriptome or specific metabolic pathway coverage.
Evolution of the annotation between Ensembl and RefSeq versions
The database contains the annotation of a probe set for different versions of Ensembl and RefSeq, which have proved to be both complementary and helpful for the interpretation of microarray gene expression data (10).
Data content of SigReannot-mart
Probes are central in the mart table structure. A probe can be linked to different probe sets (Table 2). Using alignment results, each probe will be provided with an Ensembl-gene link specificity mark which can evolve with new genome assemblies or annotations. Through the Ensembl API, based in the gene link, we fetch the orthologous genes for several species always including human and mouse as well as GO and crossreference gene identifiers.
Summary of data currently available in the SigReannot-mart database
| Microarray . | Species . | Manufacturer . | Data set . | ||
|---|---|---|---|---|---|
| Ensembl 56 . | Ensembl 59 + RefSeq RNA . | Ensembl 61+ RefSeq RNA . | |||
| 44 K | Bovine | Agilent | * | * | * |
| 24 K | EADGENE | * | |||
| 22 K | INRA | * | |||
| 44 k | Chicken | Agilent | * | * | * |
| 20 K | EADGENE | * | * | ||
| 44 K | Horse | Agilent | * | * | * |
| GPL2881 | Mouse | Agilent | * | ||
| GPL2877 | Rat | Agilent | * | ||
| 44 K | Pig | Agilent | * | * | * |
| 25 K | EADGENE | * | |||
| 17 K | INRA | * | |||
| 44 K | Rabbit | Agilent | * | * | * |
| 44 K | Salmon | Agilent | * | ||
| 15 K | Sheep | Agilent | * | * | * |
| 37 K | Trout | Agilent | * | * | |
| GPL884 | Human | Agilent | * | ||
| Microarray . | Species . | Manufacturer . | Data set . | ||
|---|---|---|---|---|---|
| Ensembl 56 . | Ensembl 59 + RefSeq RNA . | Ensembl 61+ RefSeq RNA . | |||
| 44 K | Bovine | Agilent | * | * | * |
| 24 K | EADGENE | * | |||
| 22 K | INRA | * | |||
| 44 k | Chicken | Agilent | * | * | * |
| 20 K | EADGENE | * | * | ||
| 44 K | Horse | Agilent | * | * | * |
| GPL2881 | Mouse | Agilent | * | ||
| GPL2877 | Rat | Agilent | * | ||
| 44 K | Pig | Agilent | * | * | * |
| 25 K | EADGENE | * | |||
| 17 K | INRA | * | |||
| 44 K | Rabbit | Agilent | * | * | * |
| 44 K | Salmon | Agilent | * | ||
| 15 K | Sheep | Agilent | * | * | * |
| 37 K | Trout | Agilent | * | * | |
| GPL884 | Human | Agilent | * | ||
The frequency of update of the probe set annotation follows the Ensembl update, at least two times a year. The current probe sets are not available in Ensembl.
Asterisks correspond to the annotation version of the probe sets.
Summary of data currently available in the SigReannot-mart database
| Microarray . | Species . | Manufacturer . | Data set . | ||
|---|---|---|---|---|---|
| Ensembl 56 . | Ensembl 59 + RefSeq RNA . | Ensembl 61+ RefSeq RNA . | |||
| 44 K | Bovine | Agilent | * | * | * |
| 24 K | EADGENE | * | |||
| 22 K | INRA | * | |||
| 44 k | Chicken | Agilent | * | * | * |
| 20 K | EADGENE | * | * | ||
| 44 K | Horse | Agilent | * | * | * |
| GPL2881 | Mouse | Agilent | * | ||
| GPL2877 | Rat | Agilent | * | ||
| 44 K | Pig | Agilent | * | * | * |
| 25 K | EADGENE | * | |||
| 17 K | INRA | * | |||
| 44 K | Rabbit | Agilent | * | * | * |
| 44 K | Salmon | Agilent | * | ||
| 15 K | Sheep | Agilent | * | * | * |
| 37 K | Trout | Agilent | * | * | |
| GPL884 | Human | Agilent | * | ||
| Microarray . | Species . | Manufacturer . | Data set . | ||
|---|---|---|---|---|---|
| Ensembl 56 . | Ensembl 59 + RefSeq RNA . | Ensembl 61+ RefSeq RNA . | |||
| 44 K | Bovine | Agilent | * | * | * |
| 24 K | EADGENE | * | |||
| 22 K | INRA | * | |||
| 44 k | Chicken | Agilent | * | * | * |
| 20 K | EADGENE | * | * | ||
| 44 K | Horse | Agilent | * | * | * |
| GPL2881 | Mouse | Agilent | * | ||
| GPL2877 | Rat | Agilent | * | ||
| 44 K | Pig | Agilent | * | * | * |
| 25 K | EADGENE | * | |||
| 17 K | INRA | * | |||
| 44 K | Rabbit | Agilent | * | * | * |
| 44 K | Salmon | Agilent | * | ||
| 15 K | Sheep | Agilent | * | * | * |
| 37 K | Trout | Agilent | * | * | |
| GPL884 | Human | Agilent | * | ||
The frequency of update of the probe set annotation follows the Ensembl update, at least two times a year. The current probe sets are not available in Ensembl.
Asterisks correspond to the annotation version of the probe sets.
Then, using orthologous HGNC identifiers and the KEGG files, we extract KEGG orthologs (KO) and pathways related to the probe. GO diagram or enrichment analysis can easily be performed using the text ouput formats. The HTML output format links the identifiers with the corresponding web pages (Table 3) from the KEGG, Ensembl, HGNC or Amigo web sites.
External databases referenced from SigReannot-mart
| Data source . | Genes . | Transcripts . | Pathways . | GO terms . | Gene symbols . | Orthologs . | URL . | Entities description . |
|---|---|---|---|---|---|---|---|---|
| Ensembl | * | * | * | www.ensembl.org | Gene, ncRNA, mRNA, putative RNA and orthologuous genes | |||
| RefSeq | * | http://www.ncbi.nlm.nih .gov/RefSeq/ | Transcript | |||||
| Gene Ontology | * | http://www.geneontology .org/ | GO term | |||||
| HGNC | * | http://www.genenames .org/ | Gene symbol | |||||
| KEGG | * | * | http://www.genome.jp/kegg/ | Enzyme, pathways and ortholog groups |
| Data source . | Genes . | Transcripts . | Pathways . | GO terms . | Gene symbols . | Orthologs . | URL . | Entities description . |
|---|---|---|---|---|---|---|---|---|
| Ensembl | * | * | * | www.ensembl.org | Gene, ncRNA, mRNA, putative RNA and orthologuous genes | |||
| RefSeq | * | http://www.ncbi.nlm.nih .gov/RefSeq/ | Transcript | |||||
| Gene Ontology | * | http://www.geneontology .org/ | GO term | |||||
| HGNC | * | http://www.genenames .org/ | Gene symbol | |||||
| KEGG | * | * | http://www.genome.jp/kegg/ | Enzyme, pathways and ortholog groups |
Asterisks represent the data sources corresponding to each biological entities imported in SigReannot-mart to perform the annotation process.
External databases referenced from SigReannot-mart
| Data source . | Genes . | Transcripts . | Pathways . | GO terms . | Gene symbols . | Orthologs . | URL . | Entities description . |
|---|---|---|---|---|---|---|---|---|
| Ensembl | * | * | * | www.ensembl.org | Gene, ncRNA, mRNA, putative RNA and orthologuous genes | |||
| RefSeq | * | http://www.ncbi.nlm.nih .gov/RefSeq/ | Transcript | |||||
| Gene Ontology | * | http://www.geneontology .org/ | GO term | |||||
| HGNC | * | http://www.genenames .org/ | Gene symbol | |||||
| KEGG | * | * | http://www.genome.jp/kegg/ | Enzyme, pathways and ortholog groups |
| Data source . | Genes . | Transcripts . | Pathways . | GO terms . | Gene symbols . | Orthologs . | URL . | Entities description . |
|---|---|---|---|---|---|---|---|---|
| Ensembl | * | * | * | www.ensembl.org | Gene, ncRNA, mRNA, putative RNA and orthologuous genes | |||
| RefSeq | * | http://www.ncbi.nlm.nih .gov/RefSeq/ | Transcript | |||||
| Gene Ontology | * | http://www.geneontology .org/ | GO term | |||||
| HGNC | * | http://www.genenames .org/ | Gene symbol | |||||
| KEGG | * | * | http://www.genome.jp/kegg/ | Enzyme, pathways and ortholog groups |
Asterisks represent the data sources corresponding to each biological entities imported in SigReannot-mart to perform the annotation process.
Customized BioMart environment
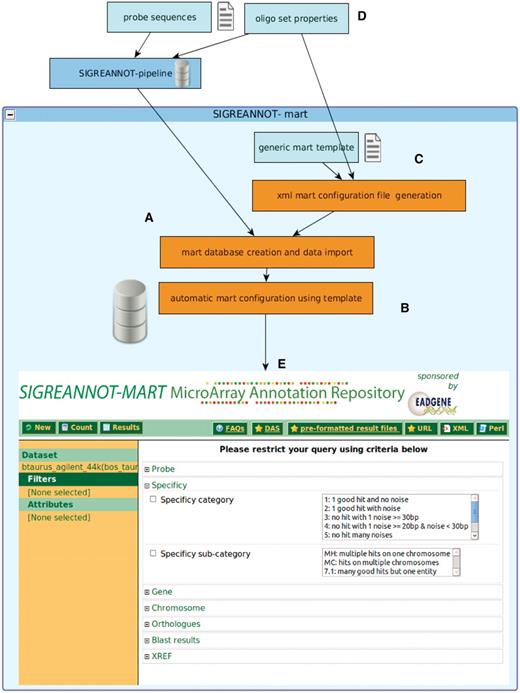
SigReannot-mart implements BioMart (11) version 0.7. For users unfamiliar with the BioMart query interface (Figure 1), pre-formatted annotations files are directly downloadable from the repository web page. For a quick overview of each data set annotation update, a summary and statistical report is also provided.
Annotation pipeline, BioMart integration and SigReannot-mart query interface. The management of the probe annotation processing pipeline and the biomart environment are centralized and automatized to allows efficient biomart configuration for multiple data sets with limited human intervention. The BioMart database is directly created and populated at the end of the annotation pipeline (A), then the BioMart configuration is automatically generated (B) using an XML file created from a generic template (C) and probe set properties (D). The SigReannot-mart data set can be filtered by user queries from a web page (E). Many attributes can be used as filters like probe specificity, Gene hits, chromosome hit location or orthologs.
A new data extraction format, the data matrix type, has been added to the BioMart Attribute page, permitting to analyze the diversity of gene categories such as KEGG pathways. This format generates a Boolean matrix indicating the membership of the probes versus pathways. This type of matrices is commonly used in R/Bioconductor (12).
Query examples
To illustrate the functions of sigReannot-mart, we present here two case studies.
Case 1: probe specificity study
A biologist wants to check if a given probe set contains probes for a list of genes he wants to monitor and how specific these probes are.
For this, he uses three criteria:
the probe set name
the gene names
the specificity (Table 1)
Step 1: select the Ensembl version in the database drop-down list.
Step 2: select the probe set in the datasets drop-down list.
Step 3: filter the probes using the IDs of the genes of interest.
Step 4: filter the probes using the specificity category 1 and 2 of the genes of interest.
BioMart query summary:
| Database | Sigenae oligo annotation |
| Dataset | btaurus_agilent_44k (bos_taurus) |
| Filters | Probe : [ID-list specified], Category: 1, 2 |
| Attributes | Probe name, gene name, specificity category |
| Database | Sigenae oligo annotation |
| Dataset | btaurus_agilent_44k (bos_taurus) |
| Filters | Probe : [ID-list specified], Category: 1, 2 |
| Attributes | Probe name, gene name, specificity category |
Analyzing the number of records representing probes for the given gene list, the biologist can decide whether or not to use this probe set.
Case 2: custom microarray design
A biologist wants to create a new probe set using existing probes and probes designed for Ensembl transcripts having no probes in the existing sets.
Step 1: Select all chicken transcript using the Ensembl BioMart.
Step 2: Select all chicken transcripts referenced by probes.
BioMart query summary:
| Database | Ensembl Gene at ensembl.org |
| Data set | Gallus gallus genes (WASHUC2) |
| Filters | |
| Attributes | Ensembl Transcript ID |
| Database | Sigenae oligo annotation at SigReannot-mart.toulouse.inra.fr |
| Datasets | ggallus_agilent_44k(sus_scrofa) and ggallus_ eadgene_20k (sus_scrofa) |
| Filters | Ensembl transcript ID ([ID-list specified) |
| Attributes | Ensembl Transcript ID |
| Database | Ensembl Gene at ensembl.org |
| Data set | Gallus gallus genes (WASHUC2) |
| Filters | |
| Attributes | Ensembl Transcript ID |
| Database | Sigenae oligo annotation at SigReannot-mart.toulouse.inra.fr |
| Datasets | ggallus_agilent_44k(sus_scrofa) and ggallus_ eadgene_20k (sus_scrofa) |
| Filters | Ensembl transcript ID ([ID-list specified) |
| Attributes | Ensembl Transcript ID |
All Ensembl transcripts found in the first query and not in the second one are not related to any probes and represent valuable target sequence to design new probes for a custom probe set design.
Discussion and future directions
While SigReannot-mart is mainly used for gene expression microarray probe set quality re-assessment and re-annotation (13), it can also be used to facilitate parts of the probe design process.
Gene expression microarrays are still widely used and therefore it is important to go on with the re-annotation process. New probe designs are still coming out and probe selection is currently going on: making these tasks easier contributes to the effort of offering accurate tools to monitor gene expression. Today, the SigReannot-mart data processing is industrialized enough for us to think about a user’s interface permitting to add a new probe set by uploading a FASTA file and indicating the corresponding species. The users would be able, a few hours later, to query the resulting annotations. Alternatively, the FASTA files of public probe sets received by email can already be processed, even if the related animal species are not currently supported (Table 4). Another even simpler option would be to schedule the annotation updates for existing probe sets. These two features should be made available in the near future.
Species related to SigReannot-mart present data sets
| Species . | Category . |
|---|---|
| Cow | Farm |
| Chicken | |
| Horse | |
| Pig | |
| Rabbit | |
| Sheep | |
| Salmon | Fishery |
| Trout | |
| Mouse | Model |
| Rat | |
| Human | |
| Species . | Category . |
|---|---|
| Cow | Farm |
| Chicken | |
| Horse | |
| Pig | |
| Rabbit | |
| Sheep | |
| Salmon | Fishery |
| Trout | |
| Mouse | Model |
| Rat | |
| Human | |
Species related to SigReannot-mart present data sets
| Species . | Category . |
|---|---|
| Cow | Farm |
| Chicken | |
| Horse | |
| Pig | |
| Rabbit | |
| Sheep | |
| Salmon | Fishery |
| Trout | |
| Mouse | Model |
| Rat | |
| Human | |
| Species . | Category . |
|---|---|
| Cow | Farm |
| Chicken | |
| Horse | |
| Pig | |
| Rabbit | |
| Sheep | |
| Salmon | Fishery |
| Trout | |
| Mouse | Model |
| Rat | |
| Human | |
Funding
EC-funded Network of Excellence EADGENE. Funding for open access charge: INRA.
Conflict of interest. None declared.