-
PDF
- Split View
-
Views
-
Cite
Cite
Rezarta Islamaj Doğan, Sun Kim, Andrew Chatr-aryamontri, Christie S. Chang, Rose Oughtred, Jennifer Rust, W. John Wilbur, Donald C. Comeau, Kara Dolinski, Mike Tyers, The BioC-BioGRID corpus: full text articles annotated for curation of protein–protein and genetic interactions, Database, Volume 2017, 2017, baw147, https://doi.org/10.1093/database/baw147
Close - Share Icon Share
Abstract
A great deal of information on the molecular genetics and biochemistry of model organisms has been reported in the scientific literature. However, this data is typically described in free text form and is not readily amenable to computational analyses. To this end, the BioGRID database systematically curates the biomedical literature for genetic and protein interaction data. This data is provided in a standardized computationally tractable format and includes structured annotation of experimental evidence. BioGRID curation necessarily involves substantial human effort by expert curators who must read each publication to extract the relevant information. Computational text-mining methods offer the potential to augment and accelerate manual curation. To facilitate the development of practical text-mining strategies, a new challenge was organized in BioCreative V for the BioC task, the collaborative Biocurator Assistant Task. This was a non-competitive, cooperative task in which the participants worked together to build BioC-compatible modules into an integrated pipeline to assist BioGRID curators. As an integral part of this task, a test collection of full text articles was developed that contained both biological entity annotations (gene/protein and organism/species) and molecular interaction annotations (protein–protein and genetic interactions (PPIs and GIs)). This collection, which we call the BioC-BioGRID corpus, was annotated by four BioGRID curators over three rounds of annotation and contains 120 full text articles curated in a dataset representing two major model organisms, namely budding yeast and human. The BioC-BioGRID corpus contains annotations for 6409 mentions of genes and their Entrez Gene IDs, 186 mentions of organism names and their NCBI Taxonomy IDs, 1867 mentions of PPIs and 701 annotations of PPI experimental evidence statements, 856 mentions of GIs and 399 annotations of GI evidence statements. The purpose, characteristics and possible future uses of the BioC-BioGRID corpus are detailed in this report.
Database URL:http://bioc.sourceforge.net/BioC-BioGRID.html
Introduction
BioCreative (Critical Assessment of Information Extraction in Biology) (1–4) is a collaborative initiative to provide a common evaluation framework for monitoring and assessing the state-of-the-art of text-mining systems applied to biologically relevant problems. The goal of the BioCreative challenges has been to pose tasks that will result in systems capable of scaling for use by general biology researchers and more specialized end users such as database curators. An important contribution of BioCreative challenges has also been the generation of shared gold standard datasets, prepared by domain experts, for the training and testing of text-mining applications. These collections, and the associated evaluation methods, represent an important resource for continued development and improvement of text-mining applications.
The BioCreative V Workshop was held in September 2015 and consisted of five tasks: the Collaborative Biocurator Assistant Task (BioC task) (5), the CHEMDNER-patents task (6), the CDR task (7), the BEL task (8, 9) and the User Interactive Task (10). Our focus is the BioC task. BioC is a format for sharing text data and annotations and a minimalistic approach to interoperability for biomedical text mining (11). The first BioC task in BioCreative IV (12) released the BioC libraries and called for development of other tools and data resources that used BioC to facilitate interoperability. The BioC task in BioCreative V was designed to make maximal use of BioC to promote data sharing and ease of use and reuse of software created. The task was positioned as a collaboration rather than a competition such that participating teams created complementary modules that could be seamlessly integrated into a system capable of assisting BioGRID curators. Specifically, the resulting interactive system triaged sentences from full text articles in order to identify text passages associated with protein–protein and genetic interactions (abbreviated PPI and GI, respectively). These sentences were then highlighted in the biocurator assistant viewer (13). Nine teams, world-wide, developed one or more modules independently (13–18), integrated via BioC, to insure the interoperability of the different systems (11).
The BioC task delivered an annotation interface that BioGRID curators then tested for their curation needs. The curation requirements addressed by the BioC annotation interface were: (i) handling full text articles, (ii) identifying molecular interactions with evidence for curation and (iii) linking genes/proteins and organisms/species to the NCBI Entrez ID and Taxonomy ID, respectively (19).
Generally, in order for a text-mining system to be of genuine use to biocurators, its performance and functionality need to be developed to optimize its usability (20). The usability of a text-mining system for biocurators requires an interactive interface that allows a variable display of annotations, links to supporting evidence and the ability to edit annotations, among other features. A key feature of such a system is how the text-mining results are presented to biocurators—i.e. what biocurators actually see in the annotation tool—and whether the text describing evidence needed for curation is highlighted.
Currently, no manually curated datasets are available that annotate only the minimal text required by the curator to capture relevant interactions and associated experimental evidence. This is in contrast with automated text-mining systems, which typically over-annotate the text. Previous efforts for similar tasks in biomedical information extraction research have produced a corpus for PPIs as a result of the BioCreative II PPI task (21), and a corpus for gene function curation via Gene Ontology (GO) annotation as a result of the BioCreative IV GO task (22). These tasks are similar in that curators were asked to annotate sentences that describe the information that was curated from the given articles. These sentences might directly describe experimental results that provided evidence for the respective annotation, or might be summary sentences that described a discovery in concise but general form. Although the GO task asked curators to mark every occurrence of GO evidence text, the PPI task asked curators to find and annotate the best evidence passage. The BioCreative V BioC task attempted to take a more natural approach by having curators mark the sentences they actually used as the basis for each interaction annotation. For all these tasks, expert manual curation represents a laborious and time consuming activity. The corpus presented here includes an important Inter-Annotator Agreement (IAA) component, which was not measured in the BioCreative II PPI task, and was reported at only 40–60% for the BioCreative IV GO task. Other corpora, such as that produced for the BioCreative II.5 challenge (23), and the one produced for BioCreative III PPI challenge (24) targeted the identifications of interacting pairs and experimental methods, respectively.
For the BioC biocurator assistant system we aimed to optimize the integration of the results of individual text-mining modules and display them in a curation interface that was most useful to the BioGRID curators. For this reason, it was necessary to generate an annotated data set of full text articles that could serve both as a gold standard to evaluate our system, and also as a benchmark for curator requirements. This effort resulted in the BioC-BioGRID corpus, which is discussed in this article.
To define an annotation task appropriate for the development of a curation tool, the organizing team collaborated with curators to develop annotation guidelines. First, BioGRID curators annotated 120 full text articles according to BioGRID curation guidelines (URL: http://wiki.thebiogrid.org/doku.php/curation_guide), and at the request of the task organizers, marked those text passages that they found useful for curation. In order to ease the annotation burden, the organizing team provided a web-based annotation interface that saved the information as marked by the curators. To provide a high-confidence gold standard set of annotations, 60 articles were annotated multiple times by different curators, which also established IAA. The final confirmation annotation round was interspersed with molecular interaction predictions generated via text-mining for curators to assess their usefulness. We assessed curator agreement amongst themselves and their agreement with text-mining predictions.
The BioC-BioGRID corpus is the first dataset, to our knowledge, that contains full text articles annotated for curation of both PPIs and GIs. It covers articles for human and yeast, and half of the corpus contains annotations from two curators as well as revisions and confirmations by two more curators. We aimed to include the sentences that are useful to a database curator for entering molecular interactions described in the article in the database, which makes this effort different from the previously curated interaction datasets.
Frequent communication between text-mining developers and curators during the course of this task identified important curation needs and highlighted ways the text-mining effort could address those needs. The BioC-BioGRID corpus can be used as gold standard training data to search for solutions that can bridge the gap between curators and text-mining algorithm developers. It consists of 6409 annotated mentions of gene/protein names associated with their Entrez Gene IDs, 186 annotated mentions of organism/species names associated with their NCBI Taxonomy IDs, 1867 annotated mentions of protein–protein annotations, 701 annotations for descriptions of their experimental methods, 856 annotated mentions of GIs and 399 annotations that mark GI evidence statements. The BioC-BioGRID corpus is a unique and valuable resource because: (i) it was produced by database curators to address their own curation needs, and (ii) it is the first corpus annotated for GI mentions and descriptions of their interaction types. We believe the BioC-BioGRID corpus should serve as an important resource for developing effective text-mining methods and as such it is available without restrictions to the community from http://bioc.sourceforge.net/BioC-BioGRID.html.
Materials and methods
Article selection
For this study, we selected a set of articles that (i) were Open-Access or for which we were granted permission from the publisher to use in this research, (ii) were curated in BioGRID, (iii) were a balanced set of both PPIs and GIs and (iv) were a balanced set for two organisms, human and the budding yeast Saccharomyces cerevisiae. We chose a representative selection of articles that reported molecular interactions in human and the budding yeast, since the ultimate goal was to enable insights across model organisms to facilitate the understanding of human disease and physiology. As yeast is amongst the best-studied organisms, and as different research communities use slightly different ways to describe similar molecular interactions, the addition of these articles provides important contextual information for text-mining purposes. Table 1 contains the distribution of the corpus articles.
Article distribution in BioC-BioGRID corpus (5)
| Organism . | PMC articles . | Interaction type . |
|---|---|---|
| Yeast | 60 | PPI and GI |
| Human | 38 | PPI and GI |
| Human | 17 | PPI |
| Human | 5 | GI |
| Organism . | PMC articles . | Interaction type . |
|---|---|---|
| Yeast | 60 | PPI and GI |
| Human | 38 | PPI and GI |
| Human | 17 | PPI |
| Human | 5 | GI |
Article distribution in BioC-BioGRID corpus (5)
| Organism . | PMC articles . | Interaction type . |
|---|---|---|
| Yeast | 60 | PPI and GI |
| Human | 38 | PPI and GI |
| Human | 17 | PPI |
| Human | 5 | GI |
| Organism . | PMC articles . | Interaction type . |
|---|---|---|
| Yeast | 60 | PPI and GI |
| Human | 38 | PPI and GI |
| Human | 17 | PPI |
| Human | 5 | GI |
Annotation task
As a general rule, curators read full text articles with the purpose of identifying curatable information that needs to be added to their particular biological database. For BioGRID, curatable information is defined as: (i) PPIs and/or GIs for organisms of interest, (ii) primary information that is not attributed to previous articles and (iii) information that is directly supported by unequivocal experimental evidence within the article. BioGRID curators were asked to manually highlight full text passages, and/or entities, which describe the interaction data that is curated.
Curators are expected to be thorough and efficient to maximize curation throughput. Contrary to what text-mining algorithm developers would normally prefer, curators do not need to identify every possible mention of every single entity of interest. Hence, their preference would be for a text-mining system that produces succinct reporting that is compatible with rapid article perusal. This aspect was made clear in our task, where we observed that the curators generally highlighted a modest number of sentences of the full text articles. In contrast, a typical text-mining system would strive to extract all mentions of the information to be added to the database.
Annotation guidelines, annotation tool and annotation data format
Annotation guidelines for the BioC-BioGRID corpus can be summarized as follows:
For each full text article, BioGRID curators curated PPIs and/or GIs described in the article, as specified by the BioGRID curation guidelines.
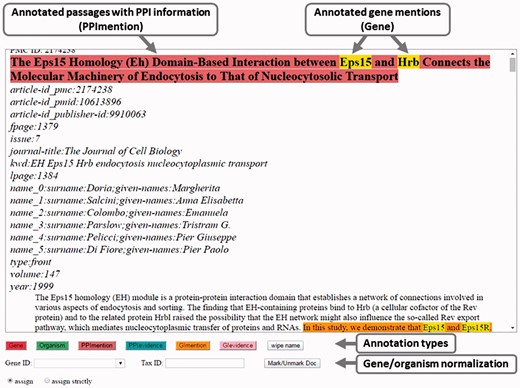
Curators used the visual interface (Figure 1) to mark the useful text passages that helped them curate the article. Useful text passages could contain mentions of the PPI or GI, or they could contain evidence in the form of important keywords that describe the experimental methods or the interaction types, which were employed by the authors.
Within these passages, curators marked the genes/proteins and their Entrez Gene IDs, and the organisms/species and their NCBI Taxonomy IDs, which were needed as identifiers for their database.
Annotation interface for the BioC-BioGRID corpus (5). Overlapping annotation types are shown in yellow in the interface. Here, gene names appear yellow because they are annotated as both ‘Gene’ and as part of a mention sentence.
Figure 1 is a screen shot of the curation tool that was built for the purpose of assisting the annotators in creating the BioC-BioGRID corpus. Curators, after selecting one of the assigned articles, had the option of scrolling through the entire full text. When reading an article using the annotation tool, curators first decided on the molecular interactions for which the current article provided evidence. Next, they highlighted supporting sentences or indicative text passages that featured those PPI or GIs. Annotations were differentiated into the actual interactions and the supporting experimental evidence. Some example text passages illustrating the kind of annotations in the BioC-BioGRID corpus are shown in Table 2. In addition to highlighting informative sentences and text passages, curators used the provided annotation tool, so that within those text passages, they annotated genes/proteins of interest and manually added their corresponding Entrez Gene IDs, and likewise for species/organisms and their NCBI Taxonomy IDs.
Some text passage examples that illustrate what annotators prefer to highlight for curation purposes. The first column shows the PMIDs. The second column lists the protein-protein and genetic interactions curated in BioGRID for these corresponding articles. And the third column shows example annotations in the BioC-BioGRID corpus that were marked by the curators for the interactions in those articles.
| PMID | BioGRID interactions | Protein-protein interaction mention examples: |
| 9700157 |
|
|
| Protein-protein interaction evidence examples: | ||
| 9700157 |
|
|
| Genetic interaction mentions examples: | ||
| 21541368 | FUS–UPF1 |
|
| Genetic interaction evidence examples: | ||
| 21541368 |
| All the FUS/TLS-specific suppressors are DNA/RNA binding proteins (Table 1, top section; and Figure 6A), including ECM32, SBP1, SKO1, and VHR1. |
| PMID | BioGRID interactions | Protein-protein interaction mention examples: |
| 9700157 |
|
|
| Protein-protein interaction evidence examples: | ||
| 9700157 |
|
|
| Genetic interaction mentions examples: | ||
| 21541368 | FUS–UPF1 |
|
| Genetic interaction evidence examples: | ||
| 21541368 |
| All the FUS/TLS-specific suppressors are DNA/RNA binding proteins (Table 1, top section; and Figure 6A), including ECM32, SBP1, SKO1, and VHR1. |
Some text passage examples that illustrate what annotators prefer to highlight for curation purposes. The first column shows the PMIDs. The second column lists the protein-protein and genetic interactions curated in BioGRID for these corresponding articles. And the third column shows example annotations in the BioC-BioGRID corpus that were marked by the curators for the interactions in those articles.
| PMID | BioGRID interactions | Protein-protein interaction mention examples: |
| 9700157 |
|
|
| Protein-protein interaction evidence examples: | ||
| 9700157 |
|
|
| Genetic interaction mentions examples: | ||
| 21541368 | FUS–UPF1 |
|
| Genetic interaction evidence examples: | ||
| 21541368 |
| All the FUS/TLS-specific suppressors are DNA/RNA binding proteins (Table 1, top section; and Figure 6A), including ECM32, SBP1, SKO1, and VHR1. |
| PMID | BioGRID interactions | Protein-protein interaction mention examples: |
| 9700157 |
|
|
| Protein-protein interaction evidence examples: | ||
| 9700157 |
|
|
| Genetic interaction mentions examples: | ||
| 21541368 | FUS–UPF1 |
|
| Genetic interaction evidence examples: | ||
| 21541368 |
| All the FUS/TLS-specific suppressors are DNA/RNA binding proteins (Table 1, top section; and Figure 6A), including ECM32, SBP1, SKO1, and VHR1. |
From the outset of the task, the collaboration between the curators and text miners was motivated by the goal of creating a resource and toolset that could assist the curators with accuracy and speed. Curators highlighted only those parts of the text that were necessary to identify and curate an interaction. The BioC-BioGRID corpus thus captures only those passages judged as necessary for the curation of that article, without extraneous text. For example, PPI sentences that mentioned interactions not supported by experimental evidence in the article were not annotated. Specific annotation cases are described in the ‘Results’ section.
Data format
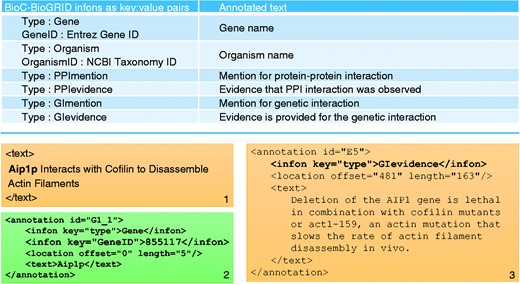
When the annotation was complete, the corpus was saved in the BioC format—a format specifically developed for sharing text data and annotations. All 120 full text articles and their annotations were thus conveniently formatted in BioC to allow for easier processing and text mining. Figure 2 summarizes the infon types used to identify different annotations in the BioC-BioGRID corpus and gives an example of an annotated text from the text mining point of view.
Summary of annotations in the BioC-BioGRID corpus. The table in the top panel lists all types of annotation infons as key:value pairs, along with a short description of what each annotation describes. The bottom panel consists of three text boxes. Text box number 1 contains an example of text from a passage in a document from the corpus. Text box number 2 shows an annotation in that passage for the gene name and its GeneID. Text box number 3 contains an annotation for a GI evidence passage.
Annotation process
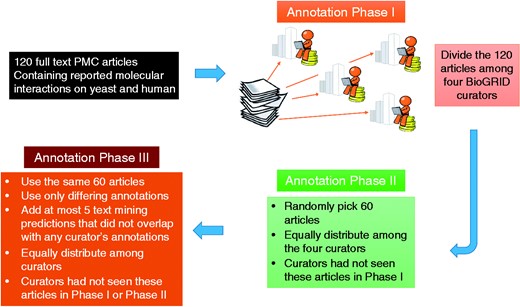
The annotation process started with the random distribution of the 120 full text articles among the four curators. There were no article overlaps. Each curator annotated 30 articles, by highlighting the relevant text that that underpinned the decision to curate one or more particular interactions. There was no limit on how many text passages the curators could mark at their discretion. All data was saved in BioC format via the annotation tool. For the second phase of annotations, 60 articles were randomly selected from the 120 articles. They were equally distributed among the same four curators so that curators were presented with articles they had not seen during Phase I. At the end of Phase II, all annotations were collected and checked for agreement. As expected, some passages overlapped, some were marked as PPI evidence by one curator while they were marked as PPI mention by the other, and some text passages did not overlap.
To better understand the usefulness of passages that were marked by only one of the curators in Phases I and II, another annotation phase was carried out, which we called the confirmation phase (Phase III). For this phase, output of text-mining tools developed for the BioC task in BioCreative V was used to (randomly) pick at most five text-mining predictions that did not overlap with any curator’s annotations. These predictions and the subset of non-overlapping annotations of Phases I and II, were combined into a new visual output for the annotation confirmation phase. This visual output presented the same 60 articles with selected pieces of text annotated for: PPI mention, PPI evidence, GI mention and/or GI evidence. Again, articles were equally distributed among curators so that each article in the 60 article set was reviewed by the two curators that had not seen the same article in the prior two phases.
There was a slight difference in the confirmation phase task compared with Phases I and II. Curators were not asked to mark the text evidence they found useful, but only to judge whether the pre-highlighted text passages were useful. It is reasonable that curators of Phases I and II could have selected different sentences supporting the same interaction. During this review phase, curators could remove all marked passages which, in their opinion, were not considered useful in curating the given article, and leave intact those that they found acceptable. We summarize the BioC-BioGRID corpus annotation process in Figure 3.
Annotation process for the BioC-BioGRID corpus. Phase I and II equally distributed the articles selected for curation among four curators so that curators had not seen the same article before. Articles contained no annotations, and curators were asked to curate them and mark the useful interactions information using the annotation interface. During Phase III, articles were equally distributed and curators were assigned articles not seen previously. Phase III articles contained pre-highlighted passages: text-mining predictions and passages annotated by only one of the Phases I or II annotators. This annotation phase asked the curators to review the annotations and remove the ones that were not useful for curation.
IAA analysis
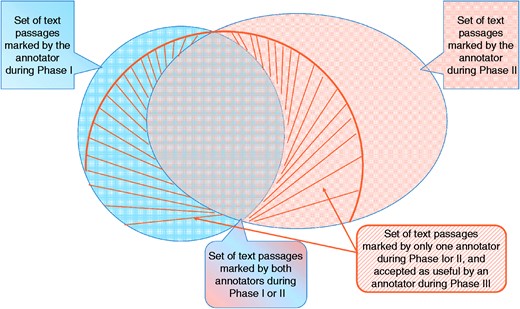
The BioC-BioGRID corpus was the product of four experienced BioGRID curators. The infographic in Figure 4 describes how we measured the corpus quality. First, Figure 4 summarizes the curators’ actions for Phases I–III. For each article, the curator reading during Phase I highlighted several text passages as useful for curation (shown in blue). A second curator reading the same article during Phase II, and unaware of what was marked during Phase I, marked a new set of text passages to help curation (shown in orange). We measure the ratio of passages that overlap over the whole set of annotations as the IAA at the end of Phase II. The passages that did not overlap with any marking of the other curator were selected for further validation and presented to the two other curators during Phase III. In this phase, the curators were presented with the full text article containing several pre-highlighted passages. The curators then decided to either keep the annotations or remove them if not useful for curation. The set of annotations accepted as useful during this phase is shown with the striped area in the Figure 4. The ratio of the agreed and accepted text passages over all annotations is reported as the IAA at the end of Phase III.
Graphic representation of IAA. For each article, an annotator highlighted several text passages as useful annotations for curation during Phase I. A second annotator reading the same article marked a different set of passages (Phase II). The two sets overlap, and also contain differences. Annotations of Phases I and II, which marked sentences that did not overlap, were re-assessed by two different curators in Phase III, where they decided whether that passage was useful or not. The striped area shows the set that was accepted during Phase III.
Analysis of text-mining predictions
As described above, Phase III of annotations used the other two curators to review the set of differing annotations from the selections of the two curators in Phases I and II. Since the curators were aware of the fact that all highlighted statements belonged to their colleagues, the organizers decided to mix in a set of text-mining predictions. These predictions were produced as a result of the BioC task in BioCreative V; however, they were randomly selected with respect to position in the article, prediction score and type of information annotated, and they were not allowed to overlap with any human annotations. While low scoring predictions would be easier for the curators to identify, the task organizers wanted to have enough of a mix to allow for some ambiguity. In addition, to avoid an unwieldy review set of annotations, the number of text-mining predictions was limited to five statements or less per annotation type. At the end of Phase III, we calculated the ratio of text-mining predictions that were accepted as useful by the curators from the whole set of the text-mining predictions that were mixed in for review. We also present the recall of the text-mining system for all the curators’ selections.
Results and discussion
Corpus overview
The BioC-BioGRID corpus resulted in 1867 annotated sentences that mark PPI mentions, 701 annotated sentences that mark PPI evidence statements, 856 annotated sentences that mark GI mentions and 399 annotated sentences that mark GI evidence statements. These numbers are summarized in Table 3 which also shows the average number of annotations and range of annotations per article in the 120 full text articles in the BioC-BioGRID corpus. Table 4 shows how the annotated statements are distributed over the two model organisms. The BioC-BioGRID corpus was selected to represent both yeast and human organisms and full text article selection aimed to have a similar coverage for them. As we see in Table 4, yeast has a larger number of articles, and a somewhat similar coverage for both GI and PPI annotations. However, we have fewer articles that contain annotations for GI evidence statements for human, due to the fact that there are currently not as many studies reporting such findings in human cells.
BioC-BioGRID corpus description of annotation types and their distribution
| Annotation type . | Range per article . | Average per article . | Number of articles . | Total . |
|---|---|---|---|---|
| PPI mention | 0–69 | 16.4 | 114 | 1867 |
| PPI evidence | 0–36 | 6.4 | 109 | 701 |
| GI mention | 0–38 | 8.8 | 97 | 856 |
| GI evidence | 0–22 | 5.3 | 76 | 399 |
| Annotation type . | Range per article . | Average per article . | Number of articles . | Total . |
|---|---|---|---|---|
| PPI mention | 0–69 | 16.4 | 114 | 1867 |
| PPI evidence | 0–36 | 6.4 | 109 | 701 |
| GI mention | 0–38 | 8.8 | 97 | 856 |
| GI evidence | 0–22 | 5.3 | 76 | 399 |
Annotation type averages are computed over the set of articles that contained that annotation type.
BioC-BioGRID corpus description of annotation types and their distribution
| Annotation type . | Range per article . | Average per article . | Number of articles . | Total . |
|---|---|---|---|---|
| PPI mention | 0–69 | 16.4 | 114 | 1867 |
| PPI evidence | 0–36 | 6.4 | 109 | 701 |
| GI mention | 0–38 | 8.8 | 97 | 856 |
| GI evidence | 0–22 | 5.3 | 76 | 399 |
| Annotation type . | Range per article . | Average per article . | Number of articles . | Total . |
|---|---|---|---|---|
| PPI mention | 0–69 | 16.4 | 114 | 1867 |
| PPI evidence | 0–36 | 6.4 | 109 | 701 |
| GI mention | 0–38 | 8.8 | 97 | 856 |
| GI evidence | 0–22 | 5.3 | 76 | 399 |
Annotation type averages are computed over the set of articles that contained that annotation type.
BioC-BioGRID corpus annotations showing the number and coverage of annotations per organism type
| Annotation type . | Number of annotations and articles (yeast) . | Number of annotations and articles (human) . |
|---|---|---|
| PPI mention | 843 (58) | 1024 (56) |
| PPI evidence | 343 (55) | 358 (54) |
| GI mention | 551 (57) | 305 (40) |
| GI evidence | 250 (49) | 149 (27) |
| Annotation type . | Number of annotations and articles (yeast) . | Number of annotations and articles (human) . |
|---|---|---|
| PPI mention | 843 (58) | 1024 (56) |
| PPI evidence | 343 (55) | 358 (54) |
| GI mention | 551 (57) | 305 (40) |
| GI evidence | 250 (49) | 149 (27) |
BioC-BioGRID corpus annotations showing the number and coverage of annotations per organism type
| Annotation type . | Number of annotations and articles (yeast) . | Number of annotations and articles (human) . |
|---|---|---|
| PPI mention | 843 (58) | 1024 (56) |
| PPI evidence | 343 (55) | 358 (54) |
| GI mention | 551 (57) | 305 (40) |
| GI evidence | 250 (49) | 149 (27) |
| Annotation type . | Number of annotations and articles (yeast) . | Number of annotations and articles (human) . |
|---|---|---|
| PPI mention | 843 (58) | 1024 (56) |
| PPI evidence | 343 (55) | 358 (54) |
| GI mention | 551 (57) | 305 (40) |
| GI evidence | 250 (49) | 149 (27) |
Inter-annotator agreement
As described earlier, experienced curators often read full text articles somewhat differently, and they may find the evidence they need for curation in different sections of a full text article. Analysing their agreement by chance (measured as the annotations’ overlap) after Phase II of annotations, we found that, generally, they mark the same section with the same annotation type 30–40% of the time. This is a much lower agreement than might have been expected for high-quality annotations. This motivated our annotation Phase III, to more accurately evaluate the quality of the data.
As described in the ‘Methods’ section, during Phase III, curators looked at articles they had not seen during Phases I or II. This time they were presented with pre-annotated portions of text, and were asked to decide whether the provided annotations were useful for curating the given article. Pre-annotations originated during Phases I or II, and represented that subset of annotations which did not overlap with any other annotation of the same article (85% of PPI mentions, 38% of PPI evidence, 61% of GI mentions and 34% of GI evidence annotations). In addition, text-mining predictions were mixed in. After Phase III, we counted the number of annotations per each type that were accepted as useful for curation and added this number to the number of overlapping annotations after Phase II. After Phase III, we saw a considerable increase in annotator agreement, as expected. In Phases I and II curators often found different sentences supporting the same interaction. Note that Phase III included only a subset of the differing annotations for review. The recomputed IAA is shown in Table 5.
Inter-annotator values measuring the overlap of annotations between Phases I and II, and how this overlap increased after Phase III was included (via checking a subset of previously non-overlapping annotations)
| Annotation type . | IAA (Phase II) . | IAA (Phase III) . |
|---|---|---|
| PPI mention | 0.38 | 0.70 |
| PPI evidence | 0.32 | 0.56 |
| GI mention | 0.42 | 0.73 |
| GI evidence | 0.40 | 0.60 |
| Annotation type . | IAA (Phase II) . | IAA (Phase III) . |
|---|---|---|
| PPI mention | 0.38 | 0.70 |
| PPI evidence | 0.32 | 0.56 |
| GI mention | 0.42 | 0.73 |
| GI evidence | 0.40 | 0.60 |
Inter-annotator values measuring the overlap of annotations between Phases I and II, and how this overlap increased after Phase III was included (via checking a subset of previously non-overlapping annotations)
| Annotation type . | IAA (Phase II) . | IAA (Phase III) . |
|---|---|---|
| PPI mention | 0.38 | 0.70 |
| PPI evidence | 0.32 | 0.56 |
| GI mention | 0.42 | 0.73 |
| GI evidence | 0.40 | 0.60 |
| Annotation type . | IAA (Phase II) . | IAA (Phase III) . |
|---|---|---|
| PPI mention | 0.38 | 0.70 |
| PPI evidence | 0.32 | 0.56 |
| GI mention | 0.42 | 0.73 |
| GI evidence | 0.40 | 0.60 |
To obtain a more granular view of the curated text, we analysed the annotations for PPI versus GI. Some annotations of Phases I and II were marked as ‘PPI mention’ (or ‘GI mention’) by one curator, and as ‘PPI evidence’ (or ‘GI evidence’) by the other, creating a perceptual mismatch (and annotator disagreement) in annotation types. Although we did not count these cases as agreement in Table 5, it is interesting to realize that, if two independent annotations on the same passage overlap, then that passage is likely to contain useful information for curation. Therefore, we calculated the annotation agreement for PPI and GI separately, and in this case, we combined mention and evidence type annotations. On the other hand, we also noticed a small number of sentences where one curator marked the text as a ‘GI mention’, and another marked the same text as ‘PPI mention’; these markings were not counted as agreement in this calculation. These results are shown in Table 6, where we clearly see a very high IAA: 88% for PPIs, and 95% for GIs.
IAA for PPI and GI passages computed as in Table 5
| Annotation type . | IAA (Phase II) . | IAA (Phase III) . |
|---|---|---|
| PPI passage | 0.54 | 0.88 |
| GI passage | 0.62 | 0.95 |
| Annotation type . | IAA (Phase II) . | IAA (Phase III) . |
|---|---|---|
| PPI passage | 0.54 | 0.88 |
| GI passage | 0.62 | 0.95 |
The mention and evidence annotations are combined for counting purposes.
IAA for PPI and GI passages computed as in Table 5
| Annotation type . | IAA (Phase II) . | IAA (Phase III) . |
|---|---|---|
| PPI passage | 0.54 | 0.88 |
| GI passage | 0.62 | 0.95 |
| Annotation type . | IAA (Phase II) . | IAA (Phase III) . |
|---|---|---|
| PPI passage | 0.54 | 0.88 |
| GI passage | 0.62 | 0.95 |
The mention and evidence annotations are combined for counting purposes.
The analysis of the text-mining predictions and their classification after the manual review, shown in Table 7, reveals two key points: First, 20–30% of text-mining predictions were in fact accepted as useful by the BioGRID curators. This is an important result and it shows that text-mining tools have considerable potential for assisting manual curation. Second, when we considered all text-mining predictions and all curators’ annotations, text-mining predictions had a recall of 70–77% of human annotations. This shows that identification of curatable molecular interaction information remains a difficult text-mining problem requiring special attention.
Curators’ selected text-mining annotations show that when presented with a random selection of text-mining predictions (that did not overlap with any curators’ annotations), the curators still find useful information
| Annotation type . | Curators’ selected text mining annotations . | Text mining recall of human annotations . |
|---|---|---|
| PPI mention | 0.26 | 0.77 |
| PPI evidence | 0.19 | 0.70 |
| GI mention/ GI evidence | 0.31 | 0.70 |
| Annotation type . | Curators’ selected text mining annotations . | Text mining recall of human annotations . |
|---|---|---|
| PPI mention | 0.26 | 0.77 |
| PPI evidence | 0.19 | 0.70 |
| GI mention/ GI evidence | 0.31 | 0.70 |
The second column shows the text mining recall of all human annotations.
Curators’ selected text-mining annotations show that when presented with a random selection of text-mining predictions (that did not overlap with any curators’ annotations), the curators still find useful information
| Annotation type . | Curators’ selected text mining annotations . | Text mining recall of human annotations . |
|---|---|---|
| PPI mention | 0.26 | 0.77 |
| PPI evidence | 0.19 | 0.70 |
| GI mention/ GI evidence | 0.31 | 0.70 |
| Annotation type . | Curators’ selected text mining annotations . | Text mining recall of human annotations . |
|---|---|---|
| PPI mention | 0.26 | 0.77 |
| PPI evidence | 0.19 | 0.70 |
| GI mention/ GI evidence | 0.31 | 0.70 |
The second column shows the text mining recall of all human annotations.
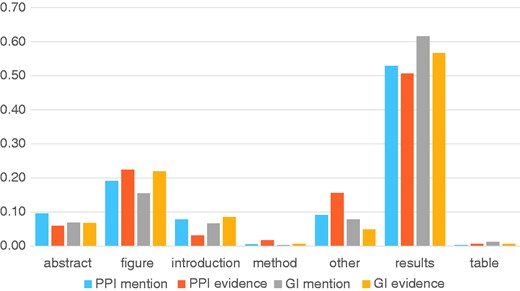
Finally, we performed an analysis of the passage titles and full text sections where the curators were more likely to annotate a PPI mention or evidence passage, or a GI mention or evidence passage. This analysis is shown in Figure 5, where we see that the ‘Results’ section is the most likely section to find molecular interaction evidence for curation, followed by ‘Figure Captions’. Fewer mentions could be found in the ‘Abstract’ or ‘Introduction’, followed by ‘Methods’ and ‘Table Captions’. A similar analysis on the text-mining predictions that were accepted as useful by the curators compared with those that were rejected also revealed a section preference. From the text-mining predictions, if the prediction was in the ‘Abstract’, ‘Results’ or ‘Figure Captions’ it was found to be useful 25–35% of the time, but much less likely if it came from a different section in the full text. These preferences correctly reflect sections where primary data for an article is reported, and suggest that automated annotation of experimental results sections might be sufficient to capture all relevant information.
Analysis of section titles of full text articles showing where different annotation types are highlighted by curators. The Y-axis shows the proportion of annotations for each annotation type.
A corpus for curation
Four BioGRID curators marked passages of interest as they read 120 full text articles to produce the BioGRID corpus. Passages of interest are those portions of text that contained evidence for the PPIs or GIs that indicate to curators that the interaction is sufficiently supported for entry into the database. Although these markings are necessary and sufficient for a human curator, they are typically considered highly ‘incomplete’ from a text-mining perspective, simply because the text annotation is not nearly exhaustive. Curator annotations do not include all mentions of PPIs or GIs that occur in a typical full text article. In particular, curator annotations exclude any mentions of PPIs or GIs which are reported in background information, are discussed in the reviewed literature or related work sections, are otherwise common knowledge, or do not describe material evidence that the interaction was observed. We also discovered that curators were careful not to include potential, unverified or hypothetical mentions of interactions. When analysing the text annotations that were rejected by the annotators during Phase III, we identified several categories that were less useful to curators. These categories are listed in Table 8. In this table, we show accordingly a selection of such cases and the associated reasons as explained during the round-table discussion of text miners and database curators.
Illustration of different sentences that are not useful for curation grouped by reasons why curators did not find them useful.
| Setup sentence |
|
| Results are not clear or description is too vague |
|
| Related literature, cited result |
|
| Interaction is described, but they are not proteins (or genes) |
| All together, our results demonstrate that the association of MUS81 with APBs is preferentially enriched at G2 phase. |
| Modification, not interaction |
|
| Setup sentence |
|
| Results are not clear or description is too vague |
|
| Related literature, cited result |
|
| Interaction is described, but they are not proteins (or genes) |
| All together, our results demonstrate that the association of MUS81 with APBs is preferentially enriched at G2 phase. |
| Modification, not interaction |
|
Illustration of different sentences that are not useful for curation grouped by reasons why curators did not find them useful.
| Setup sentence |
|
| Results are not clear or description is too vague |
|
| Related literature, cited result |
|
| Interaction is described, but they are not proteins (or genes) |
| All together, our results demonstrate that the association of MUS81 with APBs is preferentially enriched at G2 phase. |
| Modification, not interaction |
|
| Setup sentence |
|
| Results are not clear or description is too vague |
|
| Related literature, cited result |
|
| Interaction is described, but they are not proteins (or genes) |
| All together, our results demonstrate that the association of MUS81 with APBs is preferentially enriched at G2 phase. |
| Modification, not interaction |
|
Based on above analysis, our proposed guidelines for building text-mining systems that assist curation of molecular interactions would include these important parameters:
Full text processing,
Full text section analysis,
Recognition of evidence described in the current article versus information cited, referred to, or generalized from other articles,
Recognition of evidence which is supported by experimental data versus hypothetical statements, wishful or vague conclusions and inconclusive statements,
Distinction between statements describing the layout of an experiment versus the statements describing the results of that experiment,
Recognition of negative statements,
Selective display of evidence statements predicted via text-mining tools and
User-friendly design of the curation interface.
To stimulate further research into the automated annotation of biological interaction data, the BioC-BioGRID corpus is made freely available to the research community. In addition to annotations for gene/protein and organism/species entities and text passages mentioning and providing evidence for PPIs and GIs, we have added some additional information listed in Table 9. We have specifically marked all annotations produced during each annotation Phase, and we have included the detailed information collected during the review process of Phase III. We wish to provide text mining developers with information not only of the sentences that were found useful for curation, but also those that upon further review, were removed. This information is intended to provide better data for building text-mining systems that focus on curation.
Additional infons as key:value pairs that complement the BioC-BioGRID corpus
| BioC-BioGRID informs as key:value pairs . | Description of corpus annotations . |
|---|---|
| Phase_I_Annotated :1 | Produced during Phase I |
| Phase_II_Annotated :1 | Produced during Phase II |
| Phase_III_Confirmed:0 | Reviewed during Phase III and no curator found it useful |
| Phase_III_Confirmed:1 | Reviewed during Phase III and one curator found it useful |
| Phase_III_Confirmed:2 | Reviewed during Phase III and two curators found it useful |
| Text_Mining_Shown:1 | Text-mining prediction shown to curators during Phase III |
| BioC-BioGRID informs as key:value pairs . | Description of corpus annotations . |
|---|---|
| Phase_I_Annotated :1 | Produced during Phase I |
| Phase_II_Annotated :1 | Produced during Phase II |
| Phase_III_Confirmed:0 | Reviewed during Phase III and no curator found it useful |
| Phase_III_Confirmed:1 | Reviewed during Phase III and one curator found it useful |
| Phase_III_Confirmed:2 | Reviewed during Phase III and two curators found it useful |
| Text_Mining_Shown:1 | Text-mining prediction shown to curators during Phase III |
Additional infons as key:value pairs that complement the BioC-BioGRID corpus
| BioC-BioGRID informs as key:value pairs . | Description of corpus annotations . |
|---|---|
| Phase_I_Annotated :1 | Produced during Phase I |
| Phase_II_Annotated :1 | Produced during Phase II |
| Phase_III_Confirmed:0 | Reviewed during Phase III and no curator found it useful |
| Phase_III_Confirmed:1 | Reviewed during Phase III and one curator found it useful |
| Phase_III_Confirmed:2 | Reviewed during Phase III and two curators found it useful |
| Text_Mining_Shown:1 | Text-mining prediction shown to curators during Phase III |
| BioC-BioGRID informs as key:value pairs . | Description of corpus annotations . |
|---|---|
| Phase_I_Annotated :1 | Produced during Phase I |
| Phase_II_Annotated :1 | Produced during Phase II |
| Phase_III_Confirmed:0 | Reviewed during Phase III and no curator found it useful |
| Phase_III_Confirmed:1 | Reviewed during Phase III and one curator found it useful |
| Phase_III_Confirmed:2 | Reviewed during Phase III and two curators found it useful |
| Text_Mining_Shown:1 | Text-mining prediction shown to curators during Phase III |
Conclusions
Through a collaborative effort between text miners and BioGRID curators, we have generated a manually annotated corpus comprising 120 articles for both named entity recognition and molecular interaction recognition from the biomedical literature. To our knowledge, this is the first corpus of its kind annotated with a focus on the expert curation process itself. IAA results and corpus statistics verified the reliability of the corpus. Furthermore, our annotated data includes annotations for GI mention and GI evidence annotations which have not been provided as part of a manually annotated corpus before. We believe this data set will be invaluable for the development of more advanced text-mining techniques for automated extraction of biomolecular interaction data.
Funding
Intramural Research Program of the NIH, National Library of Medicine to R.I.D., S.K., W.J.W. and D.C.C.; National Institutes of Health R01OD010929 to M.T. and K.D., National Institutes of Health R24OD011194 to K.D. and M.T.
Conflict of interest. None declared.
Acknowledgements
The authors would like to thank the BioCreative V organizers and the BioCreative V BioC task participants.
References
Author notes
These authors contributed equally to this work.
Citation details: Islamaj Doğan,R., Kim,S., Chatr-Aryamontri,A. et al. The BioC-BioGRID corpus: full text articles annotated for curation of protein–protein and genetic interactions. Database (2016) Vol. 2016: article ID baw147; doi:10.1093/database/baw147