-
PDF
- Split View
-
Views
-
Cite
Cite
Kirk Roberts, Anupama E. Gururaj, Xiaoling Chen, Saeid Pournejati, William R. Hersh, Dina Demner-Fushman, Lucila Ohno-Machado, Trevor Cohen, Hua Xu, Information retrieval for biomedical datasets: the 2016 bioCADDIE dataset retrieval challenge, Database, Volume 2017, 2017, bax068, https://doi.org/10.1093/database/bax068
Close - Share Icon Share
Abstract
The focus of the 2016 bioCADDIE Dataset Retrieval Challenge was the evaluation of information retrieval techniques for identifying relevant biomedical datasets. Participants were provided with a corpus of ∼795 thousand datasets from 20 biomedical data repositories and their retrieval systems were evaluated with 15 test queries. There were 10 participants in the Challenge, submitting a total of 45 runs. The top inferred normalized discounted cumulative gain score was 0.513, while the top precision at 10 score was 0.827. The systems utilized a range of retrieval approaches, from advanced query processing to learning-to-rank frameworks. The results of the task demonstrate the potential for advanced retrieval methods in finding relevant biomedical datasets.
Introduction
Biomedical research’s increasing dependence on digital data, as well as recent concerns on experimental reproducibility and data reusability, have led to a significant increase in the number and types of datasets available to biomedical researchers. Finding data relevant to one’s own project amid the massive quantity available, however, can be quite challenging due to the diversity of information types associated with a dataset and the increasing number of sources. For this reason, new methods of information retrieval (IR) are necessary to deal with the unique challenges of dataset retrieval. To encourage the rapid development of novel methods for data discovery, a publicly available test set was created to provide a benchmark dataset on which IR researchers could compare methods. The release of this dataset was accompanied by a shared task–the 2016 bioCADDIE Dataset Retrieval Challenge–to spur rapid development and dissemination of ideas for biomedical dataset IR.
The IR challenges in searching biomedical datasets are complex. The complexity can be organized into three broad categories. First, biomedicine is fundamentally complex, both from the significant difficulties in knowledge representation and ontological reasoning and from the constant stream of new advances, ideas and paradigm shifts. In few places is this better illustrated than in the significant growth of biomedical datasets in both quantity and complexity. Second, the meta-data describing biomedical datasets combines unstructured descriptions, detailed structured data and links to scientific articles that both describe the data and leverage it for testing scientific hypotheses. All three of these data types are liable to contain large amounts of information irrelevant to the query, such as details about study authors, funding mechanisms and descriptions of prior work. Third, combining datasets from multiple repositories, while essential to a broad search strategy, results in high levels of heterogeneity of structured data, and inconsistent conventions for unstructured data. Datasets describing the results of chemical reactions, phenotype analyses and clinical trials may have extremely different data schemas. Yet individual datasets across these diverse types may still be relevant to a single query (e.g. a particular RNA-binding protein), making it critical to search broadly and reduce silo effects. Given all these complexities, and the many potential IR solutions, evaluating dataset retrieval methods on a common benchmark is vital.
In this article, we provide an overview of the 2016 bioCADDIE Dataset Retrieval Challenge, including background information that inspired the task, evaluation of submissions, results of participating systems, and a discussion on how the many innovative ideas generated during the Challenge are being integrated into the bioCADDIE project’s official search engine.
Background
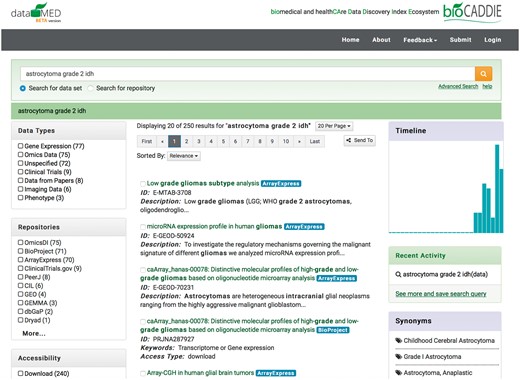
The biomedical and healthCAre Data Discovery Index Ecosystem (bioCADDIE) project (http://www.biocaddie.org) seeks to provide a prototype platform for researchers to find, reanalyze, and revise biomedical data. bioCADDIE is designed to be a common data index infrastructure, connecting with existing biomedical data repositories (e.g. dbGaP, ClinicalTrials.gov) and other data sources. The meta-data from these sources are collected, normalized and indexed. The bioCADDIE search engine—DataMed (1)—utilizes the complex and varied meta-data describing each dataset. Currently, DataMed is a relatively baseline IR system in terms of retrieval, but advanced in terms of the depth and breadth of data it ingests and indexes. DataMed indexes both structured and unstructured data from dozens of diverse dataset repositories (it is this normalized meta-data that was provided to Challenge participants). DataMed uses natural language processing to recognize biomedical concepts, then performs query expansion using the concept’s synonyms. Retrieval is performed with a simple query of concept terms with results ranked by TF-IDF. Finally, results are presented in a web interface (Figure 1 illustrates the DataMed user interface). An immediate benefit of the Dataset Retrieval Challenge is to identify innovative search strategies for integration into the DataMed search engine.
Beyond the scope of bioCADDIE and dataset retrieval, IR shared tasks have been enormously successful in fostering innovative methods and encouraging collaboration between IR researchers and biomedical experts. These shared tasks have largely been organized as part of the annual Text Retrieval Conference (TREC), organized by the US National Institute of Standards and Technology. From 2003 to 2007, the TREC Genomics track (2) focused largely on retrieving scientific articles of interest to genomics researchers. From 2011 to 2012, the TREC Medical Records track (3) switched the focus to retrieving clinical notes. Finally, from 2014 to 2016, the TREC Clinical Decision Support (CDS) track (4) focused on retrieving scientific articles of interest to clinicians. All these tasks garnered significant interest and participation, well above that of most TREC tracks. Furthermore, in support of the above point that biomedical IR shared tasks encourage collaboration between IR and biomedical researchers, we can point to the significant participation of non-biomedical researchers (largely computer and information science researchers) in the TREC tasks. For instance, in the most recent TREC CDS task (4), 18 of the 24 teams that submitted system papers were largely or entirely from non-biomedical groups. Without such shared tasks, it is highly unlikely even a small fraction of these researchers would become involved in biomedical IR since shared tasks significantly reduce the barriers to entry in the field.
The bioCADDIE Dataset Retrieval Challenge most resembles the original TREC Genomics track in its focus on researchers as the primary users. However, bioCADDIE’s focus on datasets, and not the literature, is a substantial difference for IR. Not only do datasets have their own meta-data and descriptions, they often are linked to multiple scientific articles. The focus of those articles is generally the scientific findings, and not the reusability of the data. Although a publication search engine focuses on one type of information source [scientific articles, though potentially including meta-data, e.g. Medical Subject Headings (MeSH), and citation analysis], a dataset search engine can use three very different data types: (i) structured data providing basic constraints of the dataset; (ii) textual descriptions providing a high-level overview of the data; and (iii) scientific literature and other analyses that generally are not focused on the dataset itself, but rather illustrate potential uses of the dataset. This combination of challenges has not been addressed by any previous IR shared task.
Task
The bioCADDIE Dataset Retrieval Challenge participants were provided a snapshot of the bioCADDIE index, comprising 794 992 datasets from 20 repositories in XML and JSON formats. Each dataset is accompanied by various meta-data elements. Most datasets contain some kind of description, but the other meta-data elements are repository- or dataset-specific. Table 1 shows the meta-data for three datasets (cleaner but longer versions can be seen in the Supplementary material). The first dataset is a clinical trial with mostly structured data, including its arms, inclusion criteria and funding mechanism. The second dataset is a genetic analysis of a bird species, and is relatively unstructured, with a long description and keywords. The third dataset describes the structure of a protein and is almost entirely structured data, including the individual amino acids that make up the protein. As can be seen from these three examples, the meta-data varies greatly from dataset to dataset, presenting a significant IR challenge. However, there are common structures as well (through the bioCADDIE team’s normalization efforts), so while challenging, detailed use of the structured data is possible.
Title and metadata (in JSON) of the datasets provided from the bioCADDIE index
| Title: Effect of meal replacement beverage (glucerna) on body composition, lipid metabolism and blood pressure |
| Metadata: {‘clinical_study’: {‘oversight_info’: {‘authority’: ‘China: Food and Drug Administration’, ‘has_dmc’: ‘Yes’}}, ‘dataItem’: {‘dataTypes’: [‘Study’, ‘Dataset’, ‘StudyGroup’, ‘DataSet’, ‘clinical_study’, ‘internal’, ‘Treatment’, ‘Grant’, ‘Disease’]}, ‘StudyGroup’: {‘type’: [‘Experimental’], ‘description’: [‘Glucerna 52g instead of night meal 5weeks’], ‘name’: [‘Glucerna,Meal Replacement’]}, ‘Grant’: {‘funder’: ‘Shanghai 10th People's Hospital’}, ‘Study’: {‘status’: ‘Completed’, ‘recruits’: {‘gender’: ‘Both’, ‘minimum_age’: ‘14 Years’, ‘maximum_age’: ‘65 Years’, ‘criteria’: ‘Inclusion Criteria: 1. age\uff1a14∼60years 2. BMI\u2265 28kg/m2 3. without liver, kidney, gastrointestinal and other major organic serious diseases, lactating women, pregnancy’}, ‘phase’: ‘N/A’, ‘identifier’: ‘Glucerna’, ‘homepage’: ‘https://clinicaltrials.gov/show/NCT02118389’, ‘studyType’: ‘Interventional’}, ‘Disease’: {‘name’: ‘Obesity’}, ‘DataSet’: {‘identifier’: ‘NCT02118389’}, ‘internal’: {‘link_text’: ‘Link to the current ClinicalTrials.gov record.’, ‘rank’: ‘SCR:002309’}, ‘Treatment’: {‘description’: ‘Glucerna 52g meal replacement’, ‘agent’: ‘Glucerna’, ‘title’: ‘Dietary Supplement’}, ‘Dataset’: {‘briefTitle’: ‘Effect of Meal Replacement Beverage(Glucerna) on Body Composition, Lipid Metabolism and Blood Pressure’, ‘is_fda_regulated’: ‘Yes’, ‘verificationDate’: ‘October 2013’, ‘creator’: ‘Shanghai 10th People's Hospital’, ‘title’: ‘The Effect of Meal Replacement Beverage(Glucerna) on Body Composition, Lipid Metabolism and Blood Pressure in Patients With Obesity’, ‘releaseDate’: ‘April 17, 2014’, ‘has_expanded_access’: ‘No’, ‘depositionDate’: ‘April 1, 2014’, ‘description’: ‘Effect of Meal Replacement Beverage(Glucerna) on Body Composition, lipid metabolism and blood pressure’}} |
| Title: locality info and genetic and phenotypic data for western scrub-jay museum specimens |
| Metadata: {‘datastandard’: {‘homepage’: ‘dublincore.org’, ‘name’: ‘Dublin Core’, ‘license’: ‘http://dublincore.org/about/copyright/’}, ‘dataItem’: {‘dataTypes’: [‘dataset’, ‘internal’, ‘identifiers’, ‘dataRepository’, ‘organization’, ‘datastandard’]}, ‘dataRepository’: {‘homepage’: ‘http://www.datadryad.org’, ‘ID’: ‘SCR:005910’, ‘name’: ‘Dryad Data Repository’}, ‘identifiers’: {‘ID’: [‘doi:10.5061/dryad.57f48/1’, ‘http://hdl.handle.net/10255/dryad.64554’]}, ‘dataset’: {‘dateAvailable’: ‘20140619T153456 + 0000’, ‘description’: ‘This Excel spreadsheet lists all 689 Western Scrub-Jay museum specimens used in the study. Museum abbreviations: Field Museum of Natural History (FMNH); Moore Laboratory of Zoology, Occidental College (MLZ); Museum of Vertebrate Zoology, Berkeley (MVZ). Haplotypes of the cytochrome B gene are coastal (C) or interior (I). Structure results refer to the raw results making up the different panels of Figure 3. The raw microsatellite data are listed in actual allele sizes after allele calling. For the phenotypic data: collar, vent, and eyestripe are qualitatively-scored categorical data ranging from 1 to 6. Collar is the amount of blue coming down from the shoulders under the necklace with 1 = heavy collar (coastal) and 6 = little collar (interior). Vent is the amount of blue tinging to the feathers under the tail coverts, with 1 = white feathers (coastal) and 6 = heavily blue-tinged feathers (interior). Eyestripe is the boldness of the white stripe above the eye, with 1 = bold stripe (coastal) and 6 = small stripe (interior). Other phenotypic data are in the form of continuous variables measured in millimeters. Blank cells mean there is no data taken for that individual at that variable.’, ‘license’: ‘http://creativecommons.org/publicdomain/zero/1.0/’, ‘title’: ‘Locality Info and Genetic and Phenotypic Data for Western Scrub-Jay Museum Specimens’, ‘ID’: ‘oai:datadryad.org:10255/dryad.64554’, ‘downloadURL’: ‘http://www.datadryad.org’, ‘dateIssued’: ‘20140619’, ‘keywords’: [‘birds’, ‘speciation’, ‘phylogeography’, ‘post-zygotic reproductive barriers’, ‘gene flow’], ‘dateLastUpdate’: ‘20140619’, ‘dateAccession’: ‘20140619T153456 + 0000’, ‘relatedDataset’: ‘hdl:8’}, ‘internal’: {‘recNum’: ‘99’, ‘setID’: ‘hdl_10255_2’, ‘type’: ‘Dataset’, ‘setName’: ‘Main’}, ‘organization’: {‘abbreviation’: ‘Dryad’, ‘name’: ‘Dryad Digital Repository’, ‘ID’: ‘SCR:005910’}} |
| Title: native cardosin a from cynara cardunculus L. |
| Metadata: {‘dataResource’: {‘keywords’: [], ‘altNames’: [], ‘acronyms’: []}, ‘citation’: {‘DOI’: ‘doi:10.1074/jbc.274.39.27694’, ‘author’: {‘name’: [‘Frazao, C.’, ‘Bento, I.’, ‘Costa, J.’, ‘Soares, C.M.’, ‘Verissimo, P.’, ‘Faro, C.’, ‘Pires, E.’, ‘Cooper, J.’, ‘Carrondo, M.A.’]}, ‘journal’: ‘J.Biol.Chem.’, ‘title’: ‘Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.’, ‘journalISSN’: ‘0021-9258’, ‘firstPage’: ‘27694’, ‘lastPage’: ‘27701’, ‘year’: ‘1999’, ‘PMID’: ‘pmid:10488111’}, ‘materialEntity’: [{‘formula’: ‘C2 H5 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘GLYCINE’, ‘weight’: ‘75.067’, ‘type’: ‘peptide linking’}, {‘formula’: ‘C3 H7 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘SERINE’, ‘weight’: ‘105.093’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C3 H7 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘ALANINE’, ‘weight’: ‘89.094’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H11 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘VALINE’, ‘weight’: ‘117.147’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H13 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘LEUCINE’, ‘weight’: ‘131.174’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H9 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘THREONINE’, ‘weight’: ‘119.120’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H8 N2 O3’, ‘role’: ‘chemical component’, ‘name’: ‘ASPARAGINE’, ‘weight’: ‘132.119’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H7 N O4’, ‘role’: ‘chemical component’, ‘name’: ‘ASPARTIC ACID’, ‘weight’: ‘133.104’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H15 N4 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘ARGININE’, ‘weight’: ‘175.210’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C9 H11 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘TYROSINE’, ‘weight’: ‘181.191’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C9 H11 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘PHENYLALANINE’, ‘weight’: ‘165.191’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H9 N O4’, ‘role’: ‘chemical component’, ‘name’: ‘GLUTAMIC ACID’, ‘weight’: ‘147.130’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H13 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘ISOLEUCINE’, ‘weight’: ‘131.174’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H9 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘PROLINE’, ‘weight’: ‘115.132’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H10 N2 O3’, ‘role’: ‘chemical component’, ‘name’: ‘GLUTAMINE’, ‘weight’: ‘146.146’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H15 N2 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘LYSINE’, ‘weight’: ‘147.197’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C11 H12 N2 O2’, ‘role’: ‘chemical component’, ‘name’: ‘TRYPTOPHAN’, ‘weight’: ‘204.228’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C3 H7 N O2 S’, ‘role’: ‘chemical component’, ‘name’: ‘CYSTEINE’, ‘weight’: ‘121.154’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H10 N3 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘HISTIDINE’, ‘weight’: ‘156.164’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H11 N O2 S’, ‘role’: ‘chemical component’, ‘name’: ‘METHIONINE’, ‘weight’: ‘149.207’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C8 H15 N O6’, ‘role’: ‘chemical component’, ‘name’: ‘N-ACETYL-D-GLUCOSAMINE’, ‘weight’: ‘221.210’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘C6 H12 O5’, ‘role’: ‘chemical component’, ‘name’: ‘ALPHA-L-FUCOSE’, ‘weight’: ‘164.158’, ‘type’: ‘saccharide’}, {‘formula’: ‘C6 H12 O6’, ‘role’: ‘chemical component’, ‘name’: ‘BETA-D-MANNOSE’, ‘weight’: ‘180.157’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘C6 H12 O6’, ‘role’: ‘chemical component’, ‘name’: ‘ALPHA-D-MANNOSE’, ‘weight’: ‘180.157’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘H2 O’, ‘role’: ‘chemical component’, ‘name’: ‘WATER’, ‘weight’: ‘18.015’, ‘type’: ‘non-polymer’}], ‘dataItem’: {‘keywords’: [‘HYDROLASE’, ‘HYDROLASE’, ‘ASPARTIC PROTEINASE’], ‘dataTypes’: [‘citation’, ‘materialEntity’, ‘dataItem’, ‘identifiers’], ‘title’: ‘NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L.’, ‘description’: ‘PROTEIN (CARDOSIN A) (3.4.23.-)’, ‘ID’: ‘1B5F’}, ‘identifiers’: [{‘ID’: ‘pdb:1B5F’}, {‘ID’: ‘rcsb:RCSB008008’}]} |
| Title: Effect of meal replacement beverage (glucerna) on body composition, lipid metabolism and blood pressure |
| Metadata: {‘clinical_study’: {‘oversight_info’: {‘authority’: ‘China: Food and Drug Administration’, ‘has_dmc’: ‘Yes’}}, ‘dataItem’: {‘dataTypes’: [‘Study’, ‘Dataset’, ‘StudyGroup’, ‘DataSet’, ‘clinical_study’, ‘internal’, ‘Treatment’, ‘Grant’, ‘Disease’]}, ‘StudyGroup’: {‘type’: [‘Experimental’], ‘description’: [‘Glucerna 52g instead of night meal 5weeks’], ‘name’: [‘Glucerna,Meal Replacement’]}, ‘Grant’: {‘funder’: ‘Shanghai 10th People's Hospital’}, ‘Study’: {‘status’: ‘Completed’, ‘recruits’: {‘gender’: ‘Both’, ‘minimum_age’: ‘14 Years’, ‘maximum_age’: ‘65 Years’, ‘criteria’: ‘Inclusion Criteria: 1. age\uff1a14∼60years 2. BMI\u2265 28kg/m2 3. without liver, kidney, gastrointestinal and other major organic serious diseases, lactating women, pregnancy’}, ‘phase’: ‘N/A’, ‘identifier’: ‘Glucerna’, ‘homepage’: ‘https://clinicaltrials.gov/show/NCT02118389’, ‘studyType’: ‘Interventional’}, ‘Disease’: {‘name’: ‘Obesity’}, ‘DataSet’: {‘identifier’: ‘NCT02118389’}, ‘internal’: {‘link_text’: ‘Link to the current ClinicalTrials.gov record.’, ‘rank’: ‘SCR:002309’}, ‘Treatment’: {‘description’: ‘Glucerna 52g meal replacement’, ‘agent’: ‘Glucerna’, ‘title’: ‘Dietary Supplement’}, ‘Dataset’: {‘briefTitle’: ‘Effect of Meal Replacement Beverage(Glucerna) on Body Composition, Lipid Metabolism and Blood Pressure’, ‘is_fda_regulated’: ‘Yes’, ‘verificationDate’: ‘October 2013’, ‘creator’: ‘Shanghai 10th People's Hospital’, ‘title’: ‘The Effect of Meal Replacement Beverage(Glucerna) on Body Composition, Lipid Metabolism and Blood Pressure in Patients With Obesity’, ‘releaseDate’: ‘April 17, 2014’, ‘has_expanded_access’: ‘No’, ‘depositionDate’: ‘April 1, 2014’, ‘description’: ‘Effect of Meal Replacement Beverage(Glucerna) on Body Composition, lipid metabolism and blood pressure’}} |
| Title: locality info and genetic and phenotypic data for western scrub-jay museum specimens |
| Metadata: {‘datastandard’: {‘homepage’: ‘dublincore.org’, ‘name’: ‘Dublin Core’, ‘license’: ‘http://dublincore.org/about/copyright/’}, ‘dataItem’: {‘dataTypes’: [‘dataset’, ‘internal’, ‘identifiers’, ‘dataRepository’, ‘organization’, ‘datastandard’]}, ‘dataRepository’: {‘homepage’: ‘http://www.datadryad.org’, ‘ID’: ‘SCR:005910’, ‘name’: ‘Dryad Data Repository’}, ‘identifiers’: {‘ID’: [‘doi:10.5061/dryad.57f48/1’, ‘http://hdl.handle.net/10255/dryad.64554’]}, ‘dataset’: {‘dateAvailable’: ‘20140619T153456 + 0000’, ‘description’: ‘This Excel spreadsheet lists all 689 Western Scrub-Jay museum specimens used in the study. Museum abbreviations: Field Museum of Natural History (FMNH); Moore Laboratory of Zoology, Occidental College (MLZ); Museum of Vertebrate Zoology, Berkeley (MVZ). Haplotypes of the cytochrome B gene are coastal (C) or interior (I). Structure results refer to the raw results making up the different panels of Figure 3. The raw microsatellite data are listed in actual allele sizes after allele calling. For the phenotypic data: collar, vent, and eyestripe are qualitatively-scored categorical data ranging from 1 to 6. Collar is the amount of blue coming down from the shoulders under the necklace with 1 = heavy collar (coastal) and 6 = little collar (interior). Vent is the amount of blue tinging to the feathers under the tail coverts, with 1 = white feathers (coastal) and 6 = heavily blue-tinged feathers (interior). Eyestripe is the boldness of the white stripe above the eye, with 1 = bold stripe (coastal) and 6 = small stripe (interior). Other phenotypic data are in the form of continuous variables measured in millimeters. Blank cells mean there is no data taken for that individual at that variable.’, ‘license’: ‘http://creativecommons.org/publicdomain/zero/1.0/’, ‘title’: ‘Locality Info and Genetic and Phenotypic Data for Western Scrub-Jay Museum Specimens’, ‘ID’: ‘oai:datadryad.org:10255/dryad.64554’, ‘downloadURL’: ‘http://www.datadryad.org’, ‘dateIssued’: ‘20140619’, ‘keywords’: [‘birds’, ‘speciation’, ‘phylogeography’, ‘post-zygotic reproductive barriers’, ‘gene flow’], ‘dateLastUpdate’: ‘20140619’, ‘dateAccession’: ‘20140619T153456 + 0000’, ‘relatedDataset’: ‘hdl:8’}, ‘internal’: {‘recNum’: ‘99’, ‘setID’: ‘hdl_10255_2’, ‘type’: ‘Dataset’, ‘setName’: ‘Main’}, ‘organization’: {‘abbreviation’: ‘Dryad’, ‘name’: ‘Dryad Digital Repository’, ‘ID’: ‘SCR:005910’}} |
| Title: native cardosin a from cynara cardunculus L. |
| Metadata: {‘dataResource’: {‘keywords’: [], ‘altNames’: [], ‘acronyms’: []}, ‘citation’: {‘DOI’: ‘doi:10.1074/jbc.274.39.27694’, ‘author’: {‘name’: [‘Frazao, C.’, ‘Bento, I.’, ‘Costa, J.’, ‘Soares, C.M.’, ‘Verissimo, P.’, ‘Faro, C.’, ‘Pires, E.’, ‘Cooper, J.’, ‘Carrondo, M.A.’]}, ‘journal’: ‘J.Biol.Chem.’, ‘title’: ‘Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.’, ‘journalISSN’: ‘0021-9258’, ‘firstPage’: ‘27694’, ‘lastPage’: ‘27701’, ‘year’: ‘1999’, ‘PMID’: ‘pmid:10488111’}, ‘materialEntity’: [{‘formula’: ‘C2 H5 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘GLYCINE’, ‘weight’: ‘75.067’, ‘type’: ‘peptide linking’}, {‘formula’: ‘C3 H7 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘SERINE’, ‘weight’: ‘105.093’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C3 H7 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘ALANINE’, ‘weight’: ‘89.094’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H11 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘VALINE’, ‘weight’: ‘117.147’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H13 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘LEUCINE’, ‘weight’: ‘131.174’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H9 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘THREONINE’, ‘weight’: ‘119.120’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H8 N2 O3’, ‘role’: ‘chemical component’, ‘name’: ‘ASPARAGINE’, ‘weight’: ‘132.119’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H7 N O4’, ‘role’: ‘chemical component’, ‘name’: ‘ASPARTIC ACID’, ‘weight’: ‘133.104’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H15 N4 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘ARGININE’, ‘weight’: ‘175.210’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C9 H11 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘TYROSINE’, ‘weight’: ‘181.191’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C9 H11 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘PHENYLALANINE’, ‘weight’: ‘165.191’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H9 N O4’, ‘role’: ‘chemical component’, ‘name’: ‘GLUTAMIC ACID’, ‘weight’: ‘147.130’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H13 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘ISOLEUCINE’, ‘weight’: ‘131.174’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H9 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘PROLINE’, ‘weight’: ‘115.132’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H10 N2 O3’, ‘role’: ‘chemical component’, ‘name’: ‘GLUTAMINE’, ‘weight’: ‘146.146’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H15 N2 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘LYSINE’, ‘weight’: ‘147.197’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C11 H12 N2 O2’, ‘role’: ‘chemical component’, ‘name’: ‘TRYPTOPHAN’, ‘weight’: ‘204.228’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C3 H7 N O2 S’, ‘role’: ‘chemical component’, ‘name’: ‘CYSTEINE’, ‘weight’: ‘121.154’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H10 N3 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘HISTIDINE’, ‘weight’: ‘156.164’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H11 N O2 S’, ‘role’: ‘chemical component’, ‘name’: ‘METHIONINE’, ‘weight’: ‘149.207’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C8 H15 N O6’, ‘role’: ‘chemical component’, ‘name’: ‘N-ACETYL-D-GLUCOSAMINE’, ‘weight’: ‘221.210’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘C6 H12 O5’, ‘role’: ‘chemical component’, ‘name’: ‘ALPHA-L-FUCOSE’, ‘weight’: ‘164.158’, ‘type’: ‘saccharide’}, {‘formula’: ‘C6 H12 O6’, ‘role’: ‘chemical component’, ‘name’: ‘BETA-D-MANNOSE’, ‘weight’: ‘180.157’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘C6 H12 O6’, ‘role’: ‘chemical component’, ‘name’: ‘ALPHA-D-MANNOSE’, ‘weight’: ‘180.157’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘H2 O’, ‘role’: ‘chemical component’, ‘name’: ‘WATER’, ‘weight’: ‘18.015’, ‘type’: ‘non-polymer’}], ‘dataItem’: {‘keywords’: [‘HYDROLASE’, ‘HYDROLASE’, ‘ASPARTIC PROTEINASE’], ‘dataTypes’: [‘citation’, ‘materialEntity’, ‘dataItem’, ‘identifiers’], ‘title’: ‘NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L.’, ‘description’: ‘PROTEIN (CARDOSIN A) (3.4.23.-)’, ‘ID’: ‘1B5F’}, ‘identifiers’: [{‘ID’: ‘pdb:1B5F’}, {‘ID’: ‘rcsb:RCSB008008’}]} |
The three datasets are from ClinicalTrials.gov, Dryad, and Protein Data Bank, respectively.
Title and metadata (in JSON) of the datasets provided from the bioCADDIE index
| Title: Effect of meal replacement beverage (glucerna) on body composition, lipid metabolism and blood pressure |
| Metadata: {‘clinical_study’: {‘oversight_info’: {‘authority’: ‘China: Food and Drug Administration’, ‘has_dmc’: ‘Yes’}}, ‘dataItem’: {‘dataTypes’: [‘Study’, ‘Dataset’, ‘StudyGroup’, ‘DataSet’, ‘clinical_study’, ‘internal’, ‘Treatment’, ‘Grant’, ‘Disease’]}, ‘StudyGroup’: {‘type’: [‘Experimental’], ‘description’: [‘Glucerna 52g instead of night meal 5weeks’], ‘name’: [‘Glucerna,Meal Replacement’]}, ‘Grant’: {‘funder’: ‘Shanghai 10th People's Hospital’}, ‘Study’: {‘status’: ‘Completed’, ‘recruits’: {‘gender’: ‘Both’, ‘minimum_age’: ‘14 Years’, ‘maximum_age’: ‘65 Years’, ‘criteria’: ‘Inclusion Criteria: 1. age\uff1a14∼60years 2. BMI\u2265 28kg/m2 3. without liver, kidney, gastrointestinal and other major organic serious diseases, lactating women, pregnancy’}, ‘phase’: ‘N/A’, ‘identifier’: ‘Glucerna’, ‘homepage’: ‘https://clinicaltrials.gov/show/NCT02118389’, ‘studyType’: ‘Interventional’}, ‘Disease’: {‘name’: ‘Obesity’}, ‘DataSet’: {‘identifier’: ‘NCT02118389’}, ‘internal’: {‘link_text’: ‘Link to the current ClinicalTrials.gov record.’, ‘rank’: ‘SCR:002309’}, ‘Treatment’: {‘description’: ‘Glucerna 52g meal replacement’, ‘agent’: ‘Glucerna’, ‘title’: ‘Dietary Supplement’}, ‘Dataset’: {‘briefTitle’: ‘Effect of Meal Replacement Beverage(Glucerna) on Body Composition, Lipid Metabolism and Blood Pressure’, ‘is_fda_regulated’: ‘Yes’, ‘verificationDate’: ‘October 2013’, ‘creator’: ‘Shanghai 10th People's Hospital’, ‘title’: ‘The Effect of Meal Replacement Beverage(Glucerna) on Body Composition, Lipid Metabolism and Blood Pressure in Patients With Obesity’, ‘releaseDate’: ‘April 17, 2014’, ‘has_expanded_access’: ‘No’, ‘depositionDate’: ‘April 1, 2014’, ‘description’: ‘Effect of Meal Replacement Beverage(Glucerna) on Body Composition, lipid metabolism and blood pressure’}} |
| Title: locality info and genetic and phenotypic data for western scrub-jay museum specimens |
| Metadata: {‘datastandard’: {‘homepage’: ‘dublincore.org’, ‘name’: ‘Dublin Core’, ‘license’: ‘http://dublincore.org/about/copyright/’}, ‘dataItem’: {‘dataTypes’: [‘dataset’, ‘internal’, ‘identifiers’, ‘dataRepository’, ‘organization’, ‘datastandard’]}, ‘dataRepository’: {‘homepage’: ‘http://www.datadryad.org’, ‘ID’: ‘SCR:005910’, ‘name’: ‘Dryad Data Repository’}, ‘identifiers’: {‘ID’: [‘doi:10.5061/dryad.57f48/1’, ‘http://hdl.handle.net/10255/dryad.64554’]}, ‘dataset’: {‘dateAvailable’: ‘20140619T153456 + 0000’, ‘description’: ‘This Excel spreadsheet lists all 689 Western Scrub-Jay museum specimens used in the study. Museum abbreviations: Field Museum of Natural History (FMNH); Moore Laboratory of Zoology, Occidental College (MLZ); Museum of Vertebrate Zoology, Berkeley (MVZ). Haplotypes of the cytochrome B gene are coastal (C) or interior (I). Structure results refer to the raw results making up the different panels of Figure 3. The raw microsatellite data are listed in actual allele sizes after allele calling. For the phenotypic data: collar, vent, and eyestripe are qualitatively-scored categorical data ranging from 1 to 6. Collar is the amount of blue coming down from the shoulders under the necklace with 1 = heavy collar (coastal) and 6 = little collar (interior). Vent is the amount of blue tinging to the feathers under the tail coverts, with 1 = white feathers (coastal) and 6 = heavily blue-tinged feathers (interior). Eyestripe is the boldness of the white stripe above the eye, with 1 = bold stripe (coastal) and 6 = small stripe (interior). Other phenotypic data are in the form of continuous variables measured in millimeters. Blank cells mean there is no data taken for that individual at that variable.’, ‘license’: ‘http://creativecommons.org/publicdomain/zero/1.0/’, ‘title’: ‘Locality Info and Genetic and Phenotypic Data for Western Scrub-Jay Museum Specimens’, ‘ID’: ‘oai:datadryad.org:10255/dryad.64554’, ‘downloadURL’: ‘http://www.datadryad.org’, ‘dateIssued’: ‘20140619’, ‘keywords’: [‘birds’, ‘speciation’, ‘phylogeography’, ‘post-zygotic reproductive barriers’, ‘gene flow’], ‘dateLastUpdate’: ‘20140619’, ‘dateAccession’: ‘20140619T153456 + 0000’, ‘relatedDataset’: ‘hdl:8’}, ‘internal’: {‘recNum’: ‘99’, ‘setID’: ‘hdl_10255_2’, ‘type’: ‘Dataset’, ‘setName’: ‘Main’}, ‘organization’: {‘abbreviation’: ‘Dryad’, ‘name’: ‘Dryad Digital Repository’, ‘ID’: ‘SCR:005910’}} |
| Title: native cardosin a from cynara cardunculus L. |
| Metadata: {‘dataResource’: {‘keywords’: [], ‘altNames’: [], ‘acronyms’: []}, ‘citation’: {‘DOI’: ‘doi:10.1074/jbc.274.39.27694’, ‘author’: {‘name’: [‘Frazao, C.’, ‘Bento, I.’, ‘Costa, J.’, ‘Soares, C.M.’, ‘Verissimo, P.’, ‘Faro, C.’, ‘Pires, E.’, ‘Cooper, J.’, ‘Carrondo, M.A.’]}, ‘journal’: ‘J.Biol.Chem.’, ‘title’: ‘Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.’, ‘journalISSN’: ‘0021-9258’, ‘firstPage’: ‘27694’, ‘lastPage’: ‘27701’, ‘year’: ‘1999’, ‘PMID’: ‘pmid:10488111’}, ‘materialEntity’: [{‘formula’: ‘C2 H5 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘GLYCINE’, ‘weight’: ‘75.067’, ‘type’: ‘peptide linking’}, {‘formula’: ‘C3 H7 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘SERINE’, ‘weight’: ‘105.093’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C3 H7 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘ALANINE’, ‘weight’: ‘89.094’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H11 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘VALINE’, ‘weight’: ‘117.147’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H13 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘LEUCINE’, ‘weight’: ‘131.174’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H9 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘THREONINE’, ‘weight’: ‘119.120’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H8 N2 O3’, ‘role’: ‘chemical component’, ‘name’: ‘ASPARAGINE’, ‘weight’: ‘132.119’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H7 N O4’, ‘role’: ‘chemical component’, ‘name’: ‘ASPARTIC ACID’, ‘weight’: ‘133.104’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H15 N4 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘ARGININE’, ‘weight’: ‘175.210’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C9 H11 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘TYROSINE’, ‘weight’: ‘181.191’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C9 H11 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘PHENYLALANINE’, ‘weight’: ‘165.191’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H9 N O4’, ‘role’: ‘chemical component’, ‘name’: ‘GLUTAMIC ACID’, ‘weight’: ‘147.130’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H13 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘ISOLEUCINE’, ‘weight’: ‘131.174’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H9 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘PROLINE’, ‘weight’: ‘115.132’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H10 N2 O3’, ‘role’: ‘chemical component’, ‘name’: ‘GLUTAMINE’, ‘weight’: ‘146.146’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H15 N2 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘LYSINE’, ‘weight’: ‘147.197’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C11 H12 N2 O2’, ‘role’: ‘chemical component’, ‘name’: ‘TRYPTOPHAN’, ‘weight’: ‘204.228’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C3 H7 N O2 S’, ‘role’: ‘chemical component’, ‘name’: ‘CYSTEINE’, ‘weight’: ‘121.154’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H10 N3 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘HISTIDINE’, ‘weight’: ‘156.164’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H11 N O2 S’, ‘role’: ‘chemical component’, ‘name’: ‘METHIONINE’, ‘weight’: ‘149.207’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C8 H15 N O6’, ‘role’: ‘chemical component’, ‘name’: ‘N-ACETYL-D-GLUCOSAMINE’, ‘weight’: ‘221.210’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘C6 H12 O5’, ‘role’: ‘chemical component’, ‘name’: ‘ALPHA-L-FUCOSE’, ‘weight’: ‘164.158’, ‘type’: ‘saccharide’}, {‘formula’: ‘C6 H12 O6’, ‘role’: ‘chemical component’, ‘name’: ‘BETA-D-MANNOSE’, ‘weight’: ‘180.157’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘C6 H12 O6’, ‘role’: ‘chemical component’, ‘name’: ‘ALPHA-D-MANNOSE’, ‘weight’: ‘180.157’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘H2 O’, ‘role’: ‘chemical component’, ‘name’: ‘WATER’, ‘weight’: ‘18.015’, ‘type’: ‘non-polymer’}], ‘dataItem’: {‘keywords’: [‘HYDROLASE’, ‘HYDROLASE’, ‘ASPARTIC PROTEINASE’], ‘dataTypes’: [‘citation’, ‘materialEntity’, ‘dataItem’, ‘identifiers’], ‘title’: ‘NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L.’, ‘description’: ‘PROTEIN (CARDOSIN A) (3.4.23.-)’, ‘ID’: ‘1B5F’}, ‘identifiers’: [{‘ID’: ‘pdb:1B5F’}, {‘ID’: ‘rcsb:RCSB008008’}]} |
| Title: Effect of meal replacement beverage (glucerna) on body composition, lipid metabolism and blood pressure |
| Metadata: {‘clinical_study’: {‘oversight_info’: {‘authority’: ‘China: Food and Drug Administration’, ‘has_dmc’: ‘Yes’}}, ‘dataItem’: {‘dataTypes’: [‘Study’, ‘Dataset’, ‘StudyGroup’, ‘DataSet’, ‘clinical_study’, ‘internal’, ‘Treatment’, ‘Grant’, ‘Disease’]}, ‘StudyGroup’: {‘type’: [‘Experimental’], ‘description’: [‘Glucerna 52g instead of night meal 5weeks’], ‘name’: [‘Glucerna,Meal Replacement’]}, ‘Grant’: {‘funder’: ‘Shanghai 10th People's Hospital’}, ‘Study’: {‘status’: ‘Completed’, ‘recruits’: {‘gender’: ‘Both’, ‘minimum_age’: ‘14 Years’, ‘maximum_age’: ‘65 Years’, ‘criteria’: ‘Inclusion Criteria: 1. age\uff1a14∼60years 2. BMI\u2265 28kg/m2 3. without liver, kidney, gastrointestinal and other major organic serious diseases, lactating women, pregnancy’}, ‘phase’: ‘N/A’, ‘identifier’: ‘Glucerna’, ‘homepage’: ‘https://clinicaltrials.gov/show/NCT02118389’, ‘studyType’: ‘Interventional’}, ‘Disease’: {‘name’: ‘Obesity’}, ‘DataSet’: {‘identifier’: ‘NCT02118389’}, ‘internal’: {‘link_text’: ‘Link to the current ClinicalTrials.gov record.’, ‘rank’: ‘SCR:002309’}, ‘Treatment’: {‘description’: ‘Glucerna 52g meal replacement’, ‘agent’: ‘Glucerna’, ‘title’: ‘Dietary Supplement’}, ‘Dataset’: {‘briefTitle’: ‘Effect of Meal Replacement Beverage(Glucerna) on Body Composition, Lipid Metabolism and Blood Pressure’, ‘is_fda_regulated’: ‘Yes’, ‘verificationDate’: ‘October 2013’, ‘creator’: ‘Shanghai 10th People's Hospital’, ‘title’: ‘The Effect of Meal Replacement Beverage(Glucerna) on Body Composition, Lipid Metabolism and Blood Pressure in Patients With Obesity’, ‘releaseDate’: ‘April 17, 2014’, ‘has_expanded_access’: ‘No’, ‘depositionDate’: ‘April 1, 2014’, ‘description’: ‘Effect of Meal Replacement Beverage(Glucerna) on Body Composition, lipid metabolism and blood pressure’}} |
| Title: locality info and genetic and phenotypic data for western scrub-jay museum specimens |
| Metadata: {‘datastandard’: {‘homepage’: ‘dublincore.org’, ‘name’: ‘Dublin Core’, ‘license’: ‘http://dublincore.org/about/copyright/’}, ‘dataItem’: {‘dataTypes’: [‘dataset’, ‘internal’, ‘identifiers’, ‘dataRepository’, ‘organization’, ‘datastandard’]}, ‘dataRepository’: {‘homepage’: ‘http://www.datadryad.org’, ‘ID’: ‘SCR:005910’, ‘name’: ‘Dryad Data Repository’}, ‘identifiers’: {‘ID’: [‘doi:10.5061/dryad.57f48/1’, ‘http://hdl.handle.net/10255/dryad.64554’]}, ‘dataset’: {‘dateAvailable’: ‘20140619T153456 + 0000’, ‘description’: ‘This Excel spreadsheet lists all 689 Western Scrub-Jay museum specimens used in the study. Museum abbreviations: Field Museum of Natural History (FMNH); Moore Laboratory of Zoology, Occidental College (MLZ); Museum of Vertebrate Zoology, Berkeley (MVZ). Haplotypes of the cytochrome B gene are coastal (C) or interior (I). Structure results refer to the raw results making up the different panels of Figure 3. The raw microsatellite data are listed in actual allele sizes after allele calling. For the phenotypic data: collar, vent, and eyestripe are qualitatively-scored categorical data ranging from 1 to 6. Collar is the amount of blue coming down from the shoulders under the necklace with 1 = heavy collar (coastal) and 6 = little collar (interior). Vent is the amount of blue tinging to the feathers under the tail coverts, with 1 = white feathers (coastal) and 6 = heavily blue-tinged feathers (interior). Eyestripe is the boldness of the white stripe above the eye, with 1 = bold stripe (coastal) and 6 = small stripe (interior). Other phenotypic data are in the form of continuous variables measured in millimeters. Blank cells mean there is no data taken for that individual at that variable.’, ‘license’: ‘http://creativecommons.org/publicdomain/zero/1.0/’, ‘title’: ‘Locality Info and Genetic and Phenotypic Data for Western Scrub-Jay Museum Specimens’, ‘ID’: ‘oai:datadryad.org:10255/dryad.64554’, ‘downloadURL’: ‘http://www.datadryad.org’, ‘dateIssued’: ‘20140619’, ‘keywords’: [‘birds’, ‘speciation’, ‘phylogeography’, ‘post-zygotic reproductive barriers’, ‘gene flow’], ‘dateLastUpdate’: ‘20140619’, ‘dateAccession’: ‘20140619T153456 + 0000’, ‘relatedDataset’: ‘hdl:8’}, ‘internal’: {‘recNum’: ‘99’, ‘setID’: ‘hdl_10255_2’, ‘type’: ‘Dataset’, ‘setName’: ‘Main’}, ‘organization’: {‘abbreviation’: ‘Dryad’, ‘name’: ‘Dryad Digital Repository’, ‘ID’: ‘SCR:005910’}} |
| Title: native cardosin a from cynara cardunculus L. |
| Metadata: {‘dataResource’: {‘keywords’: [], ‘altNames’: [], ‘acronyms’: []}, ‘citation’: {‘DOI’: ‘doi:10.1074/jbc.274.39.27694’, ‘author’: {‘name’: [‘Frazao, C.’, ‘Bento, I.’, ‘Costa, J.’, ‘Soares, C.M.’, ‘Verissimo, P.’, ‘Faro, C.’, ‘Pires, E.’, ‘Cooper, J.’, ‘Carrondo, M.A.’]}, ‘journal’: ‘J.Biol.Chem.’, ‘title’: ‘Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.’, ‘journalISSN’: ‘0021-9258’, ‘firstPage’: ‘27694’, ‘lastPage’: ‘27701’, ‘year’: ‘1999’, ‘PMID’: ‘pmid:10488111’}, ‘materialEntity’: [{‘formula’: ‘C2 H5 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘GLYCINE’, ‘weight’: ‘75.067’, ‘type’: ‘peptide linking’}, {‘formula’: ‘C3 H7 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘SERINE’, ‘weight’: ‘105.093’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C3 H7 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘ALANINE’, ‘weight’: ‘89.094’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H11 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘VALINE’, ‘weight’: ‘117.147’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H13 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘LEUCINE’, ‘weight’: ‘131.174’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H9 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘THREONINE’, ‘weight’: ‘119.120’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H8 N2 O3’, ‘role’: ‘chemical component’, ‘name’: ‘ASPARAGINE’, ‘weight’: ‘132.119’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C4 H7 N O4’, ‘role’: ‘chemical component’, ‘name’: ‘ASPARTIC ACID’, ‘weight’: ‘133.104’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H15 N4 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘ARGININE’, ‘weight’: ‘175.210’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C9 H11 N O3’, ‘role’: ‘chemical component’, ‘name’: ‘TYROSINE’, ‘weight’: ‘181.191’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C9 H11 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘PHENYLALANINE’, ‘weight’: ‘165.191’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H9 N O4’, ‘role’: ‘chemical component’, ‘name’: ‘GLUTAMIC ACID’, ‘weight’: ‘147.130’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H13 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘ISOLEUCINE’, ‘weight’: ‘131.174’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H9 N O2’, ‘role’: ‘chemical component’, ‘name’: ‘PROLINE’, ‘weight’: ‘115.132’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H10 N2 O3’, ‘role’: ‘chemical component’, ‘name’: ‘GLUTAMINE’, ‘weight’: ‘146.146’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H15 N2 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘LYSINE’, ‘weight’: ‘147.197’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C11 H12 N2 O2’, ‘role’: ‘chemical component’, ‘name’: ‘TRYPTOPHAN’, ‘weight’: ‘204.228’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C3 H7 N O2 S’, ‘role’: ‘chemical component’, ‘name’: ‘CYSTEINE’, ‘weight’: ‘121.154’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C6 H10 N3 O2 1’, ‘role’: ‘chemical component’, ‘name’: ‘HISTIDINE’, ‘weight’: ‘156.164’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C5 H11 N O2 S’, ‘role’: ‘chemical component’, ‘name’: ‘METHIONINE’, ‘weight’: ‘149.207’, ‘type’: ‘L-peptide linking’}, {‘formula’: ‘C8 H15 N O6’, ‘role’: ‘chemical component’, ‘name’: ‘N-ACETYL-D-GLUCOSAMINE’, ‘weight’: ‘221.210’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘C6 H12 O5’, ‘role’: ‘chemical component’, ‘name’: ‘ALPHA-L-FUCOSE’, ‘weight’: ‘164.158’, ‘type’: ‘saccharide’}, {‘formula’: ‘C6 H12 O6’, ‘role’: ‘chemical component’, ‘name’: ‘BETA-D-MANNOSE’, ‘weight’: ‘180.157’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘C6 H12 O6’, ‘role’: ‘chemical component’, ‘name’: ‘ALPHA-D-MANNOSE’, ‘weight’: ‘180.157’, ‘type’: ‘D-saccharide’}, {‘formula’: ‘H2 O’, ‘role’: ‘chemical component’, ‘name’: ‘WATER’, ‘weight’: ‘18.015’, ‘type’: ‘non-polymer’}], ‘dataItem’: {‘keywords’: [‘HYDROLASE’, ‘HYDROLASE’, ‘ASPARTIC PROTEINASE’], ‘dataTypes’: [‘citation’, ‘materialEntity’, ‘dataItem’, ‘identifiers’], ‘title’: ‘NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L.’, ‘description’: ‘PROTEIN (CARDOSIN A) (3.4.23.-)’, ‘ID’: ‘1B5F’}, ‘identifiers’: [{‘ID’: ‘pdb:1B5F’}, {‘ID’: ‘rcsb:RCSB008008’}]} |
The three datasets are from ClinicalTrials.gov, Dryad, and Protein Data Bank, respectively.
In addition to the corpus, participants were also provided with a set of six sample queries with corresponding manual judgments. These sample queries had been used to calibrate the manual judgment process (i.e. to assess and improve inter-rater agreement, as well as annotation guideline refinement), and as such were not as thoroughly annotated as the official test queries. A detailed description of the benchmark dataset, including the annotation process, is provided as a companion paper in this issue (5). Before the submission deadline, participants were provided with 15 test queries (without manual judgments). Table 2 shows four of the test queries, along with example dataset judgments for each query.
Sample of the test queries with selected relevant, partially relevant, and not relevant dataset titles
| Query 1: | ‘Find protein sequencing data related to bacterial chemotaxis across all databases’ |
| Relevant | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY. |
| Partially relevant | RNA-sequencing of mRNAs from control and CAP-D3 deficient Salmonella infected HT-29 cells |
| Not relevant | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109 |
| Query 2: | ‘Search for data of all types related to MIP-2 gene related to biliary atresia across all databases’ |
| Relevant | Comprehensive gene expression profile of extrahepatic bile ducts in mice with experimental biliary atresia |
| Partially relevant | Taqman Low Density Arrays based microRNAs expression profile of mouse extrahepatic bileducts and gallbladders during a mouse model of biliary atresia |
| Not relevant | Arginine Feeding: a Novel Strategy to Improve Protein Metabolism in Cancer and the Response to Surgery |
| Query 4: | ‘Find all data types related to inflammation during oxidative stress in human hepatic cells across all databases’ |
| Relevant | Silymarin Suppresses Cellular Inflammation By Inducing Reparative Stress Signaling |
| Partially relevant | Effect of Vitamin D3 Supplementation in Treatment of Irritable Bowel Syndrome |
| Not relevant | Effects of Sulfur Thermal Water Inhalation on Airway Oxidative Stress in COPD Patients |
| Query 14: | ‘Search for data on nerve cells in the substantia nigra in mice across all databases’ |
| Relevant | Age-mediated transcriptomic changes in adult mouse brain ventral tegmental area |
| Partially relevant | Global gene expression changes in rat retinal ganglion cells after experimental glaucoma |
| Not relevant | Study of poplar shoot apex methylome response to variations in soil water availability [sp] |
| Query 1: | ‘Find protein sequencing data related to bacterial chemotaxis across all databases’ |
| Relevant | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY. |
| Partially relevant | RNA-sequencing of mRNAs from control and CAP-D3 deficient Salmonella infected HT-29 cells |
| Not relevant | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109 |
| Query 2: | ‘Search for data of all types related to MIP-2 gene related to biliary atresia across all databases’ |
| Relevant | Comprehensive gene expression profile of extrahepatic bile ducts in mice with experimental biliary atresia |
| Partially relevant | Taqman Low Density Arrays based microRNAs expression profile of mouse extrahepatic bileducts and gallbladders during a mouse model of biliary atresia |
| Not relevant | Arginine Feeding: a Novel Strategy to Improve Protein Metabolism in Cancer and the Response to Surgery |
| Query 4: | ‘Find all data types related to inflammation during oxidative stress in human hepatic cells across all databases’ |
| Relevant | Silymarin Suppresses Cellular Inflammation By Inducing Reparative Stress Signaling |
| Partially relevant | Effect of Vitamin D3 Supplementation in Treatment of Irritable Bowel Syndrome |
| Not relevant | Effects of Sulfur Thermal Water Inhalation on Airway Oxidative Stress in COPD Patients |
| Query 14: | ‘Search for data on nerve cells in the substantia nigra in mice across all databases’ |
| Relevant | Age-mediated transcriptomic changes in adult mouse brain ventral tegmental area |
| Partially relevant | Global gene expression changes in rat retinal ganglion cells after experimental glaucoma |
| Not relevant | Study of poplar shoot apex methylome response to variations in soil water availability [sp] |
Sample of the test queries with selected relevant, partially relevant, and not relevant dataset titles
| Query 1: | ‘Find protein sequencing data related to bacterial chemotaxis across all databases’ |
| Relevant | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY. |
| Partially relevant | RNA-sequencing of mRNAs from control and CAP-D3 deficient Salmonella infected HT-29 cells |
| Not relevant | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109 |
| Query 2: | ‘Search for data of all types related to MIP-2 gene related to biliary atresia across all databases’ |
| Relevant | Comprehensive gene expression profile of extrahepatic bile ducts in mice with experimental biliary atresia |
| Partially relevant | Taqman Low Density Arrays based microRNAs expression profile of mouse extrahepatic bileducts and gallbladders during a mouse model of biliary atresia |
| Not relevant | Arginine Feeding: a Novel Strategy to Improve Protein Metabolism in Cancer and the Response to Surgery |
| Query 4: | ‘Find all data types related to inflammation during oxidative stress in human hepatic cells across all databases’ |
| Relevant | Silymarin Suppresses Cellular Inflammation By Inducing Reparative Stress Signaling |
| Partially relevant | Effect of Vitamin D3 Supplementation in Treatment of Irritable Bowel Syndrome |
| Not relevant | Effects of Sulfur Thermal Water Inhalation on Airway Oxidative Stress in COPD Patients |
| Query 14: | ‘Search for data on nerve cells in the substantia nigra in mice across all databases’ |
| Relevant | Age-mediated transcriptomic changes in adult mouse brain ventral tegmental area |
| Partially relevant | Global gene expression changes in rat retinal ganglion cells after experimental glaucoma |
| Not relevant | Study of poplar shoot apex methylome response to variations in soil water availability [sp] |
| Query 1: | ‘Find protein sequencing data related to bacterial chemotaxis across all databases’ |
| Relevant | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY. |
| Partially relevant | RNA-sequencing of mRNAs from control and CAP-D3 deficient Salmonella infected HT-29 cells |
| Not relevant | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109 |
| Query 2: | ‘Search for data of all types related to MIP-2 gene related to biliary atresia across all databases’ |
| Relevant | Comprehensive gene expression profile of extrahepatic bile ducts in mice with experimental biliary atresia |
| Partially relevant | Taqman Low Density Arrays based microRNAs expression profile of mouse extrahepatic bileducts and gallbladders during a mouse model of biliary atresia |
| Not relevant | Arginine Feeding: a Novel Strategy to Improve Protein Metabolism in Cancer and the Response to Surgery |
| Query 4: | ‘Find all data types related to inflammation during oxidative stress in human hepatic cells across all databases’ |
| Relevant | Silymarin Suppresses Cellular Inflammation By Inducing Reparative Stress Signaling |
| Partially relevant | Effect of Vitamin D3 Supplementation in Treatment of Irritable Bowel Syndrome |
| Not relevant | Effects of Sulfur Thermal Water Inhalation on Airway Oxidative Stress in COPD Patients |
| Query 14: | ‘Search for data on nerve cells in the substantia nigra in mice across all databases’ |
| Relevant | Age-mediated transcriptomic changes in adult mouse brain ventral tegmental area |
| Partially relevant | Global gene expression changes in rat retinal ganglion cells after experimental glaucoma |
| Not relevant | Study of poplar shoot apex methylome response to variations in soil water availability [sp] |
The task was conducted on a relatively short timeline: the corpus and sample queries were made available on 16 September, the test queries were made available on 14 November, and results were due on 2 December. This timeline is approximately one-half to two-thirds the amount of time provided for an equivalent TREC task.
Prior to the test query release, many teams expressed interest in the task. Ultimately, 10 teams submitted results. Table 3 lists the institutions that officially participated. Each participant was allowed to submit up to five submissions (or runs), each containing up to 1000 results per query.
Official participating teams in the Challenge, including the number of submissions (runs) for each participant
| Short name . | Institution . | No. runs . |
|---|---|---|
| BioMelb | University of Melbourne | 5 |
| Elsevier | Elsevier Limited | 5 |
| Emory | Emory University | 4 |
| HiTSZ-ICRC | Harbin Institute of Technology Shenzhen Graduate School | 5 |
| IAII_PUT | Poznan University of Technology | 1 |
| Mayo | Mayo Clinic | 5 |
| OHSU | Oregon Health and Science University | 5 |
| SIBTex | Swiss Institute of Bioinformatics | 5 |
| UCSD | University of California San Diego | 5 |
| UIUC GSIS | University of Illinois/National Data Service | 5 |
| Short name . | Institution . | No. runs . |
|---|---|---|
| BioMelb | University of Melbourne | 5 |
| Elsevier | Elsevier Limited | 5 |
| Emory | Emory University | 4 |
| HiTSZ-ICRC | Harbin Institute of Technology Shenzhen Graduate School | 5 |
| IAII_PUT | Poznan University of Technology | 1 |
| Mayo | Mayo Clinic | 5 |
| OHSU | Oregon Health and Science University | 5 |
| SIBTex | Swiss Institute of Bioinformatics | 5 |
| UCSD | University of California San Diego | 5 |
| UIUC GSIS | University of Illinois/National Data Service | 5 |
Official participating teams in the Challenge, including the number of submissions (runs) for each participant
| Short name . | Institution . | No. runs . |
|---|---|---|
| BioMelb | University of Melbourne | 5 |
| Elsevier | Elsevier Limited | 5 |
| Emory | Emory University | 4 |
| HiTSZ-ICRC | Harbin Institute of Technology Shenzhen Graduate School | 5 |
| IAII_PUT | Poznan University of Technology | 1 |
| Mayo | Mayo Clinic | 5 |
| OHSU | Oregon Health and Science University | 5 |
| SIBTex | Swiss Institute of Bioinformatics | 5 |
| UCSD | University of California San Diego | 5 |
| UIUC GSIS | University of Illinois/National Data Service | 5 |
| Short name . | Institution . | No. runs . |
|---|---|---|
| BioMelb | University of Melbourne | 5 |
| Elsevier | Elsevier Limited | 5 |
| Emory | Emory University | 4 |
| HiTSZ-ICRC | Harbin Institute of Technology Shenzhen Graduate School | 5 |
| IAII_PUT | Poznan University of Technology | 1 |
| Mayo | Mayo Clinic | 5 |
| OHSU | Oregon Health and Science University | 5 |
| SIBTex | Swiss Institute of Bioinformatics | 5 |
| UCSD | University of California San Diego | 5 |
| UIUC GSIS | University of Illinois/National Data Service | 5 |
Evaluation
Evaluation was conducted on a set of 20 184 manually annotated (judged) results. Each result was judged as definitely relevant (812, or 4% of the total), partially relevant (3069, or 15%), or not relevant (16 303, or 81%). Standard TREC procedures call for all judgments to be made after submission using a pooling process. For this Challenge, however, there was insufficient time after submission to enable a proper amount of judgments via pooling. Instead, the vast majority of judgments were made prior to submission. To aid this process, four baseline IR systems (essentially DataMed re-implemented in Lucene, Indri, Terrier and Semantic Vectors) were used to create a set of 18 417 results for manual judgment. After submission, pooling was then performed to ensure each submission had a reliable number of judgments. The top 10 results and a further 5% random sample of the top 100 results were chosen for manual judgment. Because these had heavy overlap with the pre-submission judgments, only 1767 new judgments were added. See Cohen et al. (5) for more details on the baseline systems and assessment process.
Three evaluation measures were utilized, two of which used a modified inferred measure since most of the top 1000 results for each submission did not in fact have a manual judgment. The three measures were:
- Inferred average precision (infAP): Inferred version of the standard average precision measure:where P(k) Is the precision at k and rel(k) is 1 if item k is relevant (0 otherwise).
- Inferred normalized discounted cumulative gain (infNDCG): Inferred version of NDCG. The discounted cumulative gain (DCG) is:This score is normalized by dividing by the ideal (best possible) DCG. Note that NDCG allows for any relevance score (not just 0/1). For the Challenge, not relevant datasets receive a score of 0, partially relevant a score of 1, and relevant a score of 2.
Precision at 10 (P@10): The precision of the top 10 results. Note that our pooling strategy assures the top 10 results are all judged, thus no inference is needed.
The official TREC evaluation script was used to score these measures. See Yilmaz et al. (6) for more information on how the inferred measures are calculated. The infNDCG was defined as the primary measure for the task, as it is often taken as the primary measure for TREC tasks with a three-class relevance scale. To complement infNDCG, P@10 was chosen as the secondary measure for its interpretability (10 results is often all a user actually reviews).
Results
The 10 participating teams submitted a total of 45 runs. The scores of the best runs for infAP, infNDCG and P@10 are shown in Table 4. The scores for all 45 runs are included in the online Supplementary materials. Different participants performed best in terms of infAP, infNDCG and P@10. This is not a surprise given our experience with previous IR challenges: each measure has its own strengths and biases. The correlation between the infAP and infNDCG scores is fairly strong (Spearman’s rank of 0.73), but the correlation between those and P@10 is much weaker (0.42 for infAP and 0.51 for infNDCG, both for the P@10 + partial ranks). This is not completely surprising: infAP and infNDCG measure the ‘long tail’ of results, while also giving maximum weight to the top 1 result. P@10 gives equal weight to the top 10, and nothing beyond.
Best official participant run on each metric
| Participant . | infAP . | infNDCG . | P@10 (+P) . | P@10 (−P) . |
|---|---|---|---|---|
| BioMelb | 0.2568 | 0.4017 | 0.7733 | 0.3333 |
| Elsevier | 0.3283 | 0.4368 | 0.8267 | 0.4267 |
| Emory | 0.2818 | 0.4241 | 0.7200 | 0.2667 |
| HiTSZ-ICRC | 0.2576 | 0.3850 | 0.7000 | 0.2800 |
| IAII PUT | 0.0876 | 0.3580 | 0.5333 | 0.1600 |
| Mayo | 0.1628 | 0.3933 | 0.7467 | 0.2600 |
| OHSU | 0.3193 | 0.4454 | 0.7600 | 0.3333 |
| SIBTex | 0.3664 | 0.4258 | 0.7533 | 0.3467 |
| UCSD | 0.2901 | 0.5132 | 0.7600 | 0.3333 |
| UIUC | 0.3228 | 0.4502 | 0.7133 | 0.2867 |
| Participant . | infAP . | infNDCG . | P@10 (+P) . | P@10 (−P) . |
|---|---|---|---|---|
| BioMelb | 0.2568 | 0.4017 | 0.7733 | 0.3333 |
| Elsevier | 0.3283 | 0.4368 | 0.8267 | 0.4267 |
| Emory | 0.2818 | 0.4241 | 0.7200 | 0.2667 |
| HiTSZ-ICRC | 0.2576 | 0.3850 | 0.7000 | 0.2800 |
| IAII PUT | 0.0876 | 0.3580 | 0.5333 | 0.1600 |
| Mayo | 0.1628 | 0.3933 | 0.7467 | 0.2600 |
| OHSU | 0.3193 | 0.4454 | 0.7600 | 0.3333 |
| SIBTex | 0.3664 | 0.4258 | 0.7533 | 0.3467 |
| UCSD | 0.2901 | 0.5132 | 0.7600 | 0.3333 |
| UIUC | 0.3228 | 0.4502 | 0.7133 | 0.2867 |
The best overall run on each metric is in bold. P@10 (+P) includes partially relevant results as relevant, while (−P) assumes partially relevant results are not relevant.
Best official participant run on each metric
| Participant . | infAP . | infNDCG . | P@10 (+P) . | P@10 (−P) . |
|---|---|---|---|---|
| BioMelb | 0.2568 | 0.4017 | 0.7733 | 0.3333 |
| Elsevier | 0.3283 | 0.4368 | 0.8267 | 0.4267 |
| Emory | 0.2818 | 0.4241 | 0.7200 | 0.2667 |
| HiTSZ-ICRC | 0.2576 | 0.3850 | 0.7000 | 0.2800 |
| IAII PUT | 0.0876 | 0.3580 | 0.5333 | 0.1600 |
| Mayo | 0.1628 | 0.3933 | 0.7467 | 0.2600 |
| OHSU | 0.3193 | 0.4454 | 0.7600 | 0.3333 |
| SIBTex | 0.3664 | 0.4258 | 0.7533 | 0.3467 |
| UCSD | 0.2901 | 0.5132 | 0.7600 | 0.3333 |
| UIUC | 0.3228 | 0.4502 | 0.7133 | 0.2867 |
| Participant . | infAP . | infNDCG . | P@10 (+P) . | P@10 (−P) . |
|---|---|---|---|---|
| BioMelb | 0.2568 | 0.4017 | 0.7733 | 0.3333 |
| Elsevier | 0.3283 | 0.4368 | 0.8267 | 0.4267 |
| Emory | 0.2818 | 0.4241 | 0.7200 | 0.2667 |
| HiTSZ-ICRC | 0.2576 | 0.3850 | 0.7000 | 0.2800 |
| IAII PUT | 0.0876 | 0.3580 | 0.5333 | 0.1600 |
| Mayo | 0.1628 | 0.3933 | 0.7467 | 0.2600 |
| OHSU | 0.3193 | 0.4454 | 0.7600 | 0.3333 |
| SIBTex | 0.3664 | 0.4258 | 0.7533 | 0.3467 |
| UCSD | 0.2901 | 0.5132 | 0.7600 | 0.3333 |
| UIUC | 0.3228 | 0.4502 | 0.7133 | 0.2867 |
The best overall run on each metric is in bold. P@10 (+P) includes partially relevant results as relevant, while (−P) assumes partially relevant results are not relevant.
For comparison, DataMed achieves an infNDCG of 0.2948 and P@10 (+P) of 0.4600 on the queries. This means every participant’s best submission outperformed the DataMed baseline, though some of the lower-performing runs performed worse.
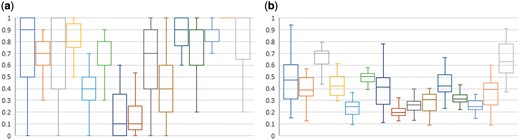
Figure 2 shows the distribution of scores by query. As can be seen, some queries are substantially more difficult than others. Query 8 (‘Search for proteomic data related to regulation of calcium in blind D. mlanogaster’) was particularly difficult, with a mean P@10 (including partial) of 0.17 and a mean infNDCG of 0.20 amongst all 45 submissions. Query 3 (‘Search for all data types related to gene TP53INP1 in relation to p53 activation across all databases’) was relatively easy, with a mean P@10 of 0.66 and a mean infNDCG of 0.64. The results for Query 14 (‘Search for data on nerve cells in the substantia nigra in mice across all databases’) were particularly strange, with a median P@10 of 1.0 (i.e. over half the submissions had a perfect top 10), a mean P@10 of 0.93 (the highest amongst the 15 queries), but a below-average mean infNDCG of 0.36. This suggests that the relevant datasets for Query 14 were almost bimodal in their retrieval difficulty: a set of at least 10 easy-to-retrieve datasets, but a substantial set of difficult-to-retrieve datasets as well.
Box-and-whiskers plot of per-query result distribution by (a) P@10, including partially relevant judgments, and (b) infNDCG.
One particularly challenging aspect of dataset retrieval is the diversity of information, in both the available meta-data and the types of data (gene expression data, clinical trial data, imaging data etc.). Within datasets from the same repository, however, there is far greater consistency in data types and meta-data. Thus it is useful to analyze results by repository. Table 5 shows the distribution of datasets, grouped by repository, in the entire DataMed index as well as the distributions for participant submissions and relevant judgments. The difference between the second (Index %) and third (Relevant %) columns in Table 5 illustrate how the test queries differed from the complete collection as a whole: datasets from the Dataverse Network Project, Dryad and Gene Expression Omnibus were far less likely to be relevant for the Challenge test queries than their distribution in the index. Meanwhile, datasets from BioProject and ArrayExpress were far more likely to be relevant to the Challenge test queries. This is not entirely surprising: the process for creating the test queries did not attempt to create a balance among the data repositories, especially in regards to their relative size, but rather followed a set of validated use cases [see Cohen et al. (5)]. An alternative hypothesis is that the baseline and participant systems used to create the judgments were particularly poor at retrieving datasets from certain repositories. The fourth column (Submit %) addresses this: it shows the distribution of datasets for the participant systems. It demonstrates that many of the datasets from less-relevant repositories were indeed retrieved, but the distribution skews closer to the relevant judgments. Finally, the differences between the third (Relevant %) and fifth (Submit Relevant %) demonstrates there was little variation between the baseline and participant systems, which is encouraging as it indicates the judged datasets are less likely to be biased toward those repositories the baseline systems tended to retrieve.
Distribution of datasets grouped by repository in (a) the DataMed index, (b) the participant submissions, (c) the complete set of datasets judged relevant for at least one query and (d) the sub-set of datasets judged relevant that were also part of a participant submission
| Repository . | Index % . | Relevant % . | Submit % . | Submit relevant % . |
|---|---|---|---|---|
| ArrayExpress | 7.7 | 22.8 | 16.4 | 23.7 |
| BioProject | 19.6 | 33.2 | 31.1 | 31.4 |
| Cancer Imaging Archive | 0.0 | 0.0 | 0.0 | 0.0 |
| CardioVascular Research Grid | 0.0 | 0.0 | 0.0 | 0.0 |
| Clinical Trials Network | 0.0 | 0.0 | 0.0 | 0.0 |
| ClinicalTrials.gov | 24.2 | 22.9 | 24.6 | 23.6 |
| Dataverse Network Project | 7.6 | 0.1 | 1.4 | 0.1 |
| Dryad Data Repository | 8.5 | 1.0 | 4.1 | 1.0 |
| GEMMA | 0.3 | 1.6 | 0.9 | 1.7 |
| Gene Expression Omnibus | 13.2 | 4.4 | 12.4 | 4.6 |
| Mouse Phenome Database | 0.0 | 0.0 | 0.1 | 0.0 |
| NeuroMorpho.Org | 4.3 | 1.1 | 0.4 | 1.2 |
| Nuclear Receptor Signaling Atlas | 0.0 | 0.0 | 0.4 | 0.0 |
| OpenfMRI | 0.0 | 0.0 | 0.0 | 0.0 |
| PeptideAtlas | 0.0 | 0.0 | 0.0 | 0.0 |
| PhenoDisco (dbGaP) | 0.1 | 0.6 | 0.2 | 0.6 |
| PhysioBank | 0.0 | 0.0 | 0.1 | 0.0 |
| ProteomeXchange | 0.2 | 0.6 | 0.5 | 0.6 |
| RCSB Protein Data Bank | 14.3 | 11.8 | 7.7 | 11.5 |
| Yale Protein Expression Database | 0.0 | 0.0 | 0.0 | 0.0 |
| Repository . | Index % . | Relevant % . | Submit % . | Submit relevant % . |
|---|---|---|---|---|
| ArrayExpress | 7.7 | 22.8 | 16.4 | 23.7 |
| BioProject | 19.6 | 33.2 | 31.1 | 31.4 |
| Cancer Imaging Archive | 0.0 | 0.0 | 0.0 | 0.0 |
| CardioVascular Research Grid | 0.0 | 0.0 | 0.0 | 0.0 |
| Clinical Trials Network | 0.0 | 0.0 | 0.0 | 0.0 |
| ClinicalTrials.gov | 24.2 | 22.9 | 24.6 | 23.6 |
| Dataverse Network Project | 7.6 | 0.1 | 1.4 | 0.1 |
| Dryad Data Repository | 8.5 | 1.0 | 4.1 | 1.0 |
| GEMMA | 0.3 | 1.6 | 0.9 | 1.7 |
| Gene Expression Omnibus | 13.2 | 4.4 | 12.4 | 4.6 |
| Mouse Phenome Database | 0.0 | 0.0 | 0.1 | 0.0 |
| NeuroMorpho.Org | 4.3 | 1.1 | 0.4 | 1.2 |
| Nuclear Receptor Signaling Atlas | 0.0 | 0.0 | 0.4 | 0.0 |
| OpenfMRI | 0.0 | 0.0 | 0.0 | 0.0 |
| PeptideAtlas | 0.0 | 0.0 | 0.0 | 0.0 |
| PhenoDisco (dbGaP) | 0.1 | 0.6 | 0.2 | 0.6 |
| PhysioBank | 0.0 | 0.0 | 0.1 | 0.0 |
| ProteomeXchange | 0.2 | 0.6 | 0.5 | 0.6 |
| RCSB Protein Data Bank | 14.3 | 11.8 | 7.7 | 11.5 |
| Yale Protein Expression Database | 0.0 | 0.0 | 0.0 | 0.0 |
Distribution of datasets grouped by repository in (a) the DataMed index, (b) the participant submissions, (c) the complete set of datasets judged relevant for at least one query and (d) the sub-set of datasets judged relevant that were also part of a participant submission
| Repository . | Index % . | Relevant % . | Submit % . | Submit relevant % . |
|---|---|---|---|---|
| ArrayExpress | 7.7 | 22.8 | 16.4 | 23.7 |
| BioProject | 19.6 | 33.2 | 31.1 | 31.4 |
| Cancer Imaging Archive | 0.0 | 0.0 | 0.0 | 0.0 |
| CardioVascular Research Grid | 0.0 | 0.0 | 0.0 | 0.0 |
| Clinical Trials Network | 0.0 | 0.0 | 0.0 | 0.0 |
| ClinicalTrials.gov | 24.2 | 22.9 | 24.6 | 23.6 |
| Dataverse Network Project | 7.6 | 0.1 | 1.4 | 0.1 |
| Dryad Data Repository | 8.5 | 1.0 | 4.1 | 1.0 |
| GEMMA | 0.3 | 1.6 | 0.9 | 1.7 |
| Gene Expression Omnibus | 13.2 | 4.4 | 12.4 | 4.6 |
| Mouse Phenome Database | 0.0 | 0.0 | 0.1 | 0.0 |
| NeuroMorpho.Org | 4.3 | 1.1 | 0.4 | 1.2 |
| Nuclear Receptor Signaling Atlas | 0.0 | 0.0 | 0.4 | 0.0 |
| OpenfMRI | 0.0 | 0.0 | 0.0 | 0.0 |
| PeptideAtlas | 0.0 | 0.0 | 0.0 | 0.0 |
| PhenoDisco (dbGaP) | 0.1 | 0.6 | 0.2 | 0.6 |
| PhysioBank | 0.0 | 0.0 | 0.1 | 0.0 |
| ProteomeXchange | 0.2 | 0.6 | 0.5 | 0.6 |
| RCSB Protein Data Bank | 14.3 | 11.8 | 7.7 | 11.5 |
| Yale Protein Expression Database | 0.0 | 0.0 | 0.0 | 0.0 |
| Repository . | Index % . | Relevant % . | Submit % . | Submit relevant % . |
|---|---|---|---|---|
| ArrayExpress | 7.7 | 22.8 | 16.4 | 23.7 |
| BioProject | 19.6 | 33.2 | 31.1 | 31.4 |
| Cancer Imaging Archive | 0.0 | 0.0 | 0.0 | 0.0 |
| CardioVascular Research Grid | 0.0 | 0.0 | 0.0 | 0.0 |
| Clinical Trials Network | 0.0 | 0.0 | 0.0 | 0.0 |
| ClinicalTrials.gov | 24.2 | 22.9 | 24.6 | 23.6 |
| Dataverse Network Project | 7.6 | 0.1 | 1.4 | 0.1 |
| Dryad Data Repository | 8.5 | 1.0 | 4.1 | 1.0 |
| GEMMA | 0.3 | 1.6 | 0.9 | 1.7 |
| Gene Expression Omnibus | 13.2 | 4.4 | 12.4 | 4.6 |
| Mouse Phenome Database | 0.0 | 0.0 | 0.1 | 0.0 |
| NeuroMorpho.Org | 4.3 | 1.1 | 0.4 | 1.2 |
| Nuclear Receptor Signaling Atlas | 0.0 | 0.0 | 0.4 | 0.0 |
| OpenfMRI | 0.0 | 0.0 | 0.0 | 0.0 |
| PeptideAtlas | 0.0 | 0.0 | 0.0 | 0.0 |
| PhenoDisco (dbGaP) | 0.1 | 0.6 | 0.2 | 0.6 |
| PhysioBank | 0.0 | 0.0 | 0.1 | 0.0 |
| ProteomeXchange | 0.2 | 0.6 | 0.5 | 0.6 |
| RCSB Protein Data Bank | 14.3 | 11.8 | 7.7 | 11.5 |
| Yale Protein Expression Database | 0.0 | 0.0 | 0.0 | 0.0 |
The approaches taken by the 10 participants varied greatly (as was hoped given the motivations of the Challenge). Many systems employed query parsers to identify terms in the query that could be matched to dataset meta-data. Most systems utilized some form of query expansion, though the knowledge sources harvested and query expansion techniques ranged widely. A common knowledge source was the US National Library of Medicine’s MeSH. Finally, some systems employed machine learning-based ranking frameworks, such as learning-to-rank, though it isn’t clear whether the quantity and quality of the data from the six sample queries was sufficient to maximize the usefulness of this technique. As such, we expect such techniques to improve dataset retrieval in the future, as more time and data are available for system improvement.
Discussion
The 2016 bioCADDIE Dataset Retrieval Challenge is, to our knowledge, the first shared task of its kind for IR of biomedical datasets. Its level of participation—10 teams—was quite positive for a shared task conducted on a short timeline (announced early September 2016, results due by 2 December). While participation was lower than that of biomedical TREC tasks, it still compares favorably to many established TREC tasks despite the longer TREC schedule. This reinforces the interest of the IR community in applying IR to biomedical tasks: these tasks tend to combine a high level of data-type complexity with an important real-world application. Beyond simply affirming the interest in biomedical IR, the bioCADDIE Dataset Challenge provided an important function in three areas of interest.
First, the challenge submissions themselves were notable. The empirical results discussed above only illustrate part of the success of these systems. With only six partially annotated queries for system development, it is likely that systems were either under- or over-tuned on these queries. Thus the scores from Table 4 likely indicate the lower bounds of performance once they are re-calibrated on the results of the 15 test queries (see discussion on scores from other biomedical IR tasks below).
Second, these 15 test queries, with over twenty thousand manual judgments, now form a publicly available benchmark for dataset retrieval. The TREC tracks have shown the value in public benchmarks: they drive further system development by IR researchers for years to come. Indeed, despite being a decade old, at least half a dozen articles were published using the TREC Genomics data in 2016 alone (7–12). It is our hope that the bioCADDIE Dataset Challenge data will form a similarly useful dataset, both for ad hoc IR tasks as well as other, as-yet-unintended uses (e.g. the TREC Genomics data is often used for similarity tasks (10) instead of just retrieval). Relative to the other biomedical IR tasks, the participant results are favorable for this task. For context, the mean and median infNDCG of the best submission for each participant for this Challenge was 0.423 and 0.425. The similar averages to the 2012 TREC Medical Records track (3) were higher (mean: 0.546, median: 0.525), but the 2015 CDS track (13) were far lower (mean: 0.229; median: 0.210), and that was the highest results of the 3 years of the CDS track. Therefore, assuming users of dataset retrieval systems have similar levels of tolerance for irrelevant results as other biomedical IR tasks, the participating systems clearly fall within an acceptable range for a usable IR system. Admittedly, not only are better retrieval systems still desired, but an understanding of the needs of dataset retrieval users is needed, in part to determine their level of tolerance for irrelevant results. The DataMed team is in the process of conducting a usability study to determine this particular need as well as other needs of dataset retrieval users. Providing a benchmark dataset, then, is important not only to evaluate systems, but also to evaluate user experiences and to encourage future contributions to research in this area. Finally, an important lesson learned from TREC is that scores on the first instance of a track are low, but improve greatly as researchers are given sufficient time and data to tune methods to the particular issues of the task (e.g. the 2014 CDS track had mean of 0.151 vs. the 2015 mean of 0.229). This suggests that one can expect higher scores on this benchmark (and dataset retrieval in general) in the future.
Third, an immediate benefit of the Challenge is that many of the innovative ideas proposed by the participants are currently being integrated into the DataMed system. These include: query expansion with neural word embeddings, learning-to-rank with query-dataset similarity features, repository weighting/prediction models and UniProt classification of both queries and datasets. The ultimate goal for DataMed is to be a PubMed for biomedical datasets, both in terms of the breadth of functionality and the significance to the community. The Challenge has thus provided the DataMed team with a wide set of ideas and experimental results upon which to determine the most promising methods for incorporation.
Finally, some thought to how future shared tasks in dataset retrieval should proceed is warranted based on the lessons learned in the Challenge. First, the most requested need in all such tasks is access to more annotated data with which to tune and train retrieval systems. This is now available as a result of this Challenge. Second, the ongoing DataMed usability study might reveal more appropriate metrics for dataset retrieval (e.g. do users only look at the top few results, or is the ‘long tail’ important to evaluate?). Third, and perhaps most ambitious, dataset queries encode complex relationships between data types, so ideally a dataset retrieval engine should indicate how a given dataset is relevant to the query. For example, given Query 4 (‘Find all data types related to inflammation during oxidative stress in human hepatic cells across all databases’), a dataset may address inflammation in human hepatic cells in general (not just oxidative stress), or a dataset may address the phenomenon in some other type of human cell (or a non-human cell). Although the ideal dataset would be a perfect match for the query, this is often not possible and, furthermore, results that aren’t exact matches may still be of use to the researcher (but probably only certain types of partial matches based on the researcher’s needs). Adding this functionality would require both a significant change in the assessment process as well as important changes in the retrieval systems. It would enable, however, a fundamentally new mechanism through which researcher can discover relevant datasets by showing the user what aspects of the query are well-covered and where gaps exist in scientific data.
Conclusion
The bioCADDIE Dataset Retrieval Challenge has not only generated benchmark data for a complex task but also engaged several teams in developing innovative solutions to the problem of finding relevant datasets across a variety of data sources. The bioCADDIE team has engaged some of the high performing teams to integrate their open source code into the DataMed search engine, which is also an open source initiative sponsored by the bioCADDIE project. We anticipate this to be the first of a series of dataset retrieval challenges and expect it to be as successful and impactful as the TREC challenges, which served as an inspiration and model for the bioCADDIE Dataset Retrieval Challenge. The data science community has much to benefit from active engagement in the development of new algorithms and tools for dataset retrieval, as well as from the availability of annotated data for their evaluation. Finding relevant data is the first step towards their integration and reuse for new discoveries. We thank all groups who participated for their contributions to this important component of biomedical data science.
Supplementary data
Supplementary data are available at Database Online.
Funding
This project was supported by the National Institutes of Health grant (U24AI117966).
Conflict of interest. None declared.
References
Author notes
Citation details: Roberts,K., Gururaj,A.E., Chen,X. et al. Information retrieval for biomedical datasets: the 2016 bioCADDIE dataset retrieval challenge. Database (2017) Vol. 2017: article ID bax068; doi:10.1093/database/bax068