-
PDF
- Split View
-
Views
-
Cite
Cite
Anuradha Surendra, Miroslava Cuperlovic-Culf, Database of resistance related metabolites in Wheat Fusarium head blight Disease (MWFD), Database, Volume 2017, 2017, bax076, https://doi.org/10.1093/database/bax076
Close - Share Icon Share
Abstract
Fungal diseases are an increasing threat to worldwide food security. Fusarium head blight (FHB), primarily caused by Fusarium graminearum, is a devastating disease of Triticum aestivum (bread wheat). Partial resistance to FHB of several wheat and barley cultivars includes specific metabolic responses to inoculation. Investigation of metabolic changes in plants, following pathogen infection, provides valuable data for understanding of the role of metabolites and metabolism in plant-pathogen interaction and resistance. Determination of functions of metabolites in resistance can also inspire the development of antifungals. Metabolic changes induced by FHB in resistant and susceptible plants have been previously investigated. However, the functionality of the majority of these investigated metabolites remains unknown. The ‘Metabolites in the Wheat Fusarium head blight Disease’ (MWFD) database was compiled in order to determine possible targets and roles of these molecules in resistance to FBH and aid in the development of related synthetic antifungals. The MWFD database allows for the quick retrieval of known resistance related metabolites, associated target proteins and their sequence analogues in wheat and Fusarium genomes. The database can be searched for compounds, MeSH terms, as well as protein targets. This comprehensive, manually curated, collection of resistance related metabolites is available at https://bioinfo.nrc.ca/mwfd/index.php.
Database URL:https://bioinfo.nrc.ca/mwfd/index.php
Introduction
Fungi cause some of the major plant ailments with approximately 11 000 plant diseases directly attributed to fungi (1). Fusarium Head Blight (FHB), primarily caused by Fusarium graminearum, is one of the most devastating diseases in wheat and barley with increasing prevalence due to changing climate, agricultural practices and suboptimal growth locations (2–6). FHB causes major losses in crop yield due to poor plant health and contamination of grains with potent mycotoxins, primarily deoxynivalenol (DON) (7, 8) and related trichothecene. Although fully resistant wheat and barley varieties are still not available, some cultivars show partial disease resistance to FHB (9). Partial resistance currently present in some plant varieties is provided through the plant immune system which includes: pathogen recognition mechanisms, signalling and transcriptional activation of defence-related genes. Activation of defence-related genes leads to cell wall reinforcement, production of anti-microbial peptides and biosynthesis of low-molecular-weight compounds (metabolites involved in the restriction of infection progression) (10, 11). These, mostly secondary metabolites (such as hormones and, phenolic and polyphenolic compounds), have long been suggested as major contributors in plant defence (12).
Several general databases of natural products have been recently established including very large assemblies such as ‘Super Natural II’ with 325 508 natural compounds with their 2 D structures, physicochemical properties, related molecules, predicted toxicity class and possible vendors (40). Several other, smaller databases of natural products have been recently reviewed in Harvey et al. (41) all providing assembly of natural products from diverse natural sources aimed to assist in drug discovery. These databases show general characteristics of natural products, purchase vendors, and in some cases; similar compounds from the complete compound space and pathways from KEGG or related resource. MWFD specifically focuses of metabolites (natural product compounds) implicated in resistance to FHB, a major threat to food production.
Understanding the function of resistance related metabolites can further the advancement in the breeding FHB resistant plants and design of novel, targeted and environmentally benign antifungals. The role of secondary metabolites in the interaction between cereal plants and pathogens has been extensively reviewed (for recent review see Ref. 13). Several metabolomics investigations [using nuclear magnetic resonance (NMR) and Mass Spectometry (MS)] have reported upon the metabolome response of resistant and susceptible wheat and barley cultivars following FHB infection. Recently, Karre et al. (14) have shown a hierarchy of regulatory genes and their downstream effects, on resistance related metabolites within the resistant barley genotype CI9831, which has been previously characterized with chitin elicitor receptor kinase HvCERK1 gene expression. They identified several resistance related metabolites belonging to phenylpropanoids, hydroxycinnamic acids and jasmonic acid pathways. Using some of the most resistant wheat lines, Dhokane et al. (15) determined a high abundance of metabolites belonging to: phenylpropanoid, lignin, glycerophospholipid, flavonoid, fatty acid and terpenoid pathways. This finding was corroborated with observed up-regulation of related genes. A number of earlier publications also provided information about resistance related metabolites in wheat and barley using MS (16–23). In addition to their role in plant pathogen response, many of these compounds are active natural products that have been subjected to variety of bioassays including cell and enzymatic testing. Based on these tests, a number of resistance related metabolites included in the dataset have proven activity against specific protein targets in different, primarily, non-fungal cells (such as Human and Mouse). Information about the activity of these compounds against any target, regardless of the biological system, is highly valuable for the determination of their role in plant-pathogen interactions. Outlining possible protein targets in fungus or wheat based on sequence similarity, to the previously revealed targets from other species, can help in determining significant targets for FHB treatment either through the development of optimized antifungal agents or breeding wheat that produces relevant resistance related metabolites. The metabolites presented in the website, were determined to be present at higher concentration in wheat and barley varieties showing some level of resistance to FHB following exposure to FHB(termed ‘over-concentrated’ in the text). The known targets for these metabolites and their homologous proteins in wheat (Triticum aestivum) and Fusarium graminearum, related species Fusarium oxysporum, are included in the database and can be searched in the website. Similarities between all resistances related metabolites are provided, allowing users to quickly determine related compounds and their possible targets in wheat and Fusarium. The database includes metabolites determined as resistance related, with possible role in plant resistance response. Resistance indicator metabolites (metabolites related to mechanisms of resistance), such as deoxynivalenol-3-O-glucoside (D3G) (18) were not included in the database.
Materials and methods
Data sources, integration and the web interface
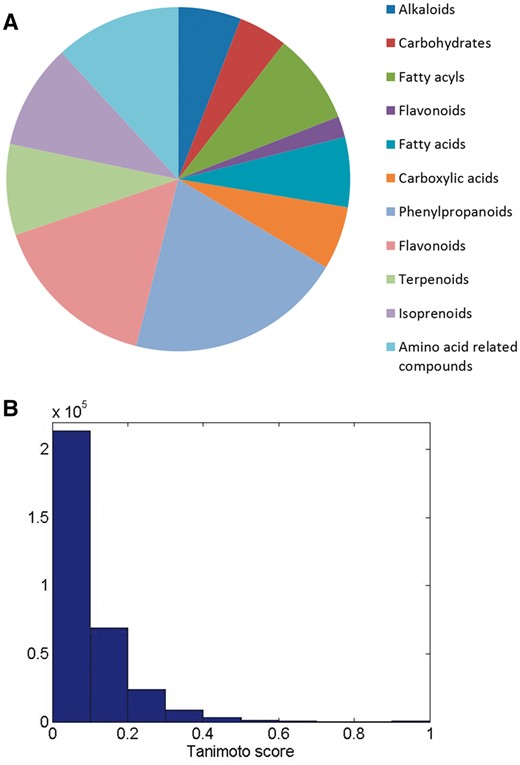
Up-regulated metabolites in plants demonstrating some level of resistance to FHB in wheat (15, 19–21, 23–27), barley (17, 18, 22), chickpeas (28) and associated as general resistance metabolites (29, 30), plant hormones (31, 32), signalling molecules (33), have been complied into the MWFD database. The MWFD database currently contains 567 metabolites, representing a diverse range of chemical structural classes (Figure 1A). Structural diversity of the metabolites in the database were assessed by Tanimoto score (Figure 1B), which is a measure of the structural similarity between molecules based on molecular fingerprints. The Tanimoto score was calculated as c/(a + b)-c, where c is the number of common molecular fragments in two compounds (compound A and compound B) and a + b is the sum of the overall number of fragments in compound A and Compound B (44).
(A) Classification of resistance related metabolites included in the database into various chemical groups. Pie chart shows relative number of compounds in each class. (B) Histogram of Tanimoto scores shows overall cross section of the level of similarities between metabolites.
Cheminformatics (such as compound structural similarity) and drug-like characteristics (such as compliance to Lipinski’s rules for drug-like compounds), of these compounds have been previously explored (34). The MWFD website is comprised of a large database of resistance related metabolites presented in a Web searchable format. The MWFD database includes the information about compound properties (45, 46) and reported inhibitory activities of these compounds against known targets (47). Although our primary interest is in the activity of metabolites against Fusarium or wheat proteins, we have provided information regarding all biological activities observed against enzymatic and whole cell targets, including mammalian cell lines. The most similar proteins to experimentally tested protein targets in Triticum aestivum and Fusarium oxysporum have been determined using BLAST sequence comparison and are presented in the database. Chemical characteristics of compounds are calculated using R routines and packages (42, 43) These characteristics can be used to find related metabolites in the database. This Web searchable database can be utilized in a variety of ways with content and search options presented in Figure 1 and several examples of use shown in this publication.
MWFD includes a user-friendly and largely self-explanatory web interface that provides access to all the data in the database. From the front page the user can browse through all the compounds in the database. Compound list shows all MWFD metabolites with their characteristics including: PubChem CID, IUPAC Name, Chemical Names, Molecular Formula, Molecular Weight, InChiKey, Canonical SMILES and Isomeric SMILES and can be sorted or searched by any of these properties. Users can access compound information directly by clicking on the PubChem CID which leads to the page on MWFD site showing all compound information as well as structure similarities with other compounds in the database and known activities of the compound. Compound searches can be done individually or in a batch and the search function is not case-sensitive. Subsets of active targets and metabolites is presented in a histogram that is developed using the R graph packages and the package Plotly (48) and provides both number of active compounds, their description, associated metabolites and a hyperlink to more information about the protein target.
Implementation
MWFD uses web-based HTML interactive interfaces combined with PHP and javascript. All data is stored in the MySQL environment and all calculations were done using R. The Database can be searched by metabolite characteristics such as: PubChem CID, compound name (including aliases and synonyms) and structure (including SMILES and InChiKey). Finally, the database can be searched by target using either a protein symbol or name. Types of searches and possible applications of MWFD are shown below.
Results
MWFD can be searched for individual metabolites or for a batch of metabolites. Individual analysis of MWFD content can be performed for compound names, CID and structures as well as target names. Specific examples of application and proposed uses for the MWFD are shown. Database content, search terms and deliverables are schematically presented in Figure 2.
Schematic description of the database content, search terms and deliverables either for the list of compounds or for single compound. Information for SINGLE COMPOUND is divided into sections listed in the table and sections marked with * can be further searched.
Examples of application of MWFD database
Target search
MWFD provides information for the testing of resistance related compounds against 867 distinct targets. Of the 567 metabolites in the database, activity against 264 proteins is proven. Target proteins can be searched using their complete or UniProt name as well as parts of the name. A ‘target protein’ search returns all compounds that have been tested previously for activity against the protein and includes inactive, active and inconclusive results. Subset of active targets can be viewed in the histogram provided for the metabolite list or in the list for an individual metabolite. The active target histogram, indicates a count of the number of metabolites associated with each active gene. The user can mouse-over each individual bar within the histogram to get the gene description, list of metabolites associated with the gene, and a link to the individual gene target search. An example of this type of search is demonstrated here using metabolites showing activity against Small Ubiquitin-like Modifier (SUMO) protein. Recent work has shown that binding of SUMO protein to other cellular proteins (SUMOylation) plays a major role in a plants’ susceptibility to necrotrophic fungal infection (35). Specifically, plants with compromised SUMOylation showed an increased susceptibility to necrotrophic fungal infection by Botrytis cinerea and Plectosphaerella cucumerina.
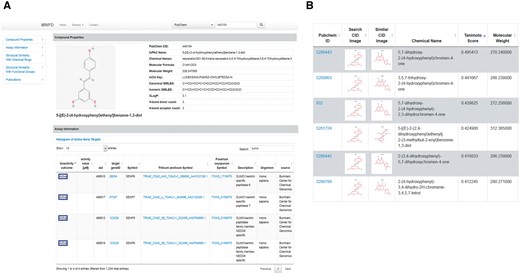
Searching by the keyword SUMO (case insensitive) under ‘Target protein’, results in three metabolites with tested activity against SUMO related targets. The effect of each metabolite can be explored in greater detail by selecting its link or by viewing and further exploring all known active targets, shown in the downloadable histogram. Benzoic acid shows several active targets. However, a subsequent search of ‘sumo’ under its targets (‘Assay Information’ section) shows that it is inactive against all 4 tested sumo related proteins. Similarly, galanthamine is shown to be inactive against SENP8, the only sumo-related protein tested. The search result for resveratrol is shown in Figure 3A. Resveratrol has been shown to be an active inhibitor of four members of SUMO/sentrin protease family in Human cells. All four of the SUMO/sentrin protease family members have paralogues (highly similar matches to proteins) in wheat. SUMO/sentrin proteases are involved in protein desumoylation (36). Thus, inhibition of SUMO/sentrin protease is hypothesized to lead to increased resistance to fungal infection (35). MWFD also shows several structurally similar compounds amongst the resistance related metabolites that were not tested against SUMOylation regulators (Figure 3B). Therefore, analysis of information provided by MWFD suggests that further investigation into SUMOylation and deSUMOylation, and the role of resveratrol and related compounds in the process of wheat resistance to Fusarium graminearum is highly warranted. This provides compounds that require further testing as inhibitors of SUMO proteases.
(A) Activity of resveratrol (CID 445154) tested against SUMOylation related proteins in human cells. MWFD shows result of tests as well as the most similar proteins in wheat and Fusarium for each determined protein target. (B) Six compounds with highest similarity to resveratrol according to Tanimoto score.
Active molecule development
The MWFD database can be searched for specific molecule or for a group of related molecules, using PubChem CID, compound name or InChiKey and SMILES. The database can also be used to annotate LC-MS data by searching m/z peaks belonging to a user defined range. Also, the database can be searched for specific functional groups by searching for keywords in the name or partial SMILES structures. The following is an example of this type of search using a phenol moiety. Several publications have recently demonstrated the antifungal activity of thymol (37, 38). MWFD can be used to investigate presence of related molecules amongst known resistance related metabolites to further the investigation of their hypothetical function. There are several different ways to search the database for thymol related molecules. Thymol is a phenol obtained from thyme oil or other volatile oils. A search under ‘compound names’ or ‘MeSH’ of ‘phenol’ reveals 22 resistance related metabolites with phenol mentioned under name including MeSH and Depositor-Supplied synonyms. The histogram of active compounds, provided as a link for the resulting list of metabolites, shows that the largest number of compounds have activity against CA2. The user can then get more information about the active molecules by mousing over the histogram columns. The MWFD can also be searched by compound structure. Structural information provided by SMILES can either be searched by exact structure or for related structures by adding a wildcard symbol (*) in the search. Thus, a search of compounds with the exact phenol group structure *C1 = CC = C(C = C1)O* leads to 6 compounds and a search for: *C1 = CC = C*C = C1*O* lists 32 resistance related metabolites. Further analysis of this subset of metabolites leads to the compound 2,6-ditert-butyl-4-methylphenol (CID 31404), with a highly similar structure to that of thymol and an associated activity as an inhibitor of human CA2 proteins. This analysis suggests that antifungal characteristics of this compound should be further investigated.
Target analysis for a group of compounds
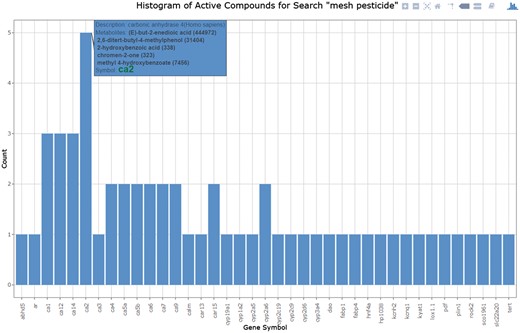
Performing a ‘compound_name’ search allows for a search of names or keywords including all PubChem listed MeSH and depositor-supplied synonyms. As an example, we can search for all compounds in the database with a known pesticide function, by searching for the keyword ‘pesticide’ under ‘compound_ name’. This search results in a list of 23 compounds, which can be further individually investigated or exported as a tab-delimited file. In addition, a histogram of active gene targets (Figure 4) can be generated to demonstrate the distribution and frequency of pesticides with associated target activities. From the histogram, known pesticide activities are associated with metaboloenzymes with the largest number of listed pesticides showing activity against Carbonic anhydrases (CA), in particular, Carbonic anhydrase 2 (CA2) in human cells. CAs are ubiquitous zinc-containing metaloenzymes that catalyse interconversion between CO2 and HCO3. CAs, thereby regulate carboxylation reactions and pH homeostasis (39). Significance of CA in fungal growth and pathogenesis has been recently studied in great detail, showing the importance of CA for fungal growth and adaptation to changing CO2 levels experienced during pathogenesis (39).
Histogram of gene counts for all known targets where tested compound or group of compounds showed activity. The active target histogram, indicates the number of metabolites associated with each active gene. The user can mouse-over each individual bar within the histogram to get the gene description, list of metabolites associated with the gene (with name and CID), and a link to the individual gene target search.
Conclusions and perspective
The MWFD database provides a unique, manually curated, assembly of metabolites that were previously shown as present during the resistance response of plants against FHB. The MWFB is a web searchable database that provides information about known protein targets for these molecules and their paralogues in wheat and Fusarium species. MWFD is the first comprehensive Web-based resource providing information about metabolites involved in resistance response to FHB and their characteristics. Our aim is for MWFD to assist in the development of more resistant wheat varieties and contribute to the focused antifungal compound discovery. In addition, similarity searches provided by MWFD show only similarities between resistances related metabolites. This allows for focused analysis of their targets and prevalent metabolic classes involved in the resistance response. MWFD also uniquely allows for a search of plant derived resistance related metabolites based on their targets and functions, which was obtained from previously published tests. Finally, the shown protein targets of resistance related metabolites, allow for further investigation of possible functions of metabolites in the resistance response while also providing promising targets for the development of antifungal agents. Some examples of MWFD use are shown in this publication.
Availability
The MWFD Database is available at: https://bioinfo.nrc.ca/mwfd/index.php. Access is fully open to academic, commercial or government based users.
Acknowledgement
Authors would like to thank Rene Richard, Eric Joyce and Chad Matsalla for their help and support that made this work possible.
Funding
This work was supported by the Canadian Wheat Alliance and National Research Council of Canada.
Conflict of interest. None declared.
References
Author notes
Citation details: Surendra,A. and Cuperlovic-Culf,M. Database of resistance related metabolites in wheat Fusarium head blight disease (MWFD). Database (2017) Vol. 2017: article ID bax076; doi:10.1093/database/bax076