-
PDF
- Split View
-
Views
-
Cite
Cite
Rezarta Islamaj, W John Wilbur, Natalie Xie, Noreen R Gonzales, Narmada Thanki, Roxanne Yamashita, Chanjuan Zheng, Aron Marchler-Bauer, Zhiyong Lu, PubMed Text Similarity Model and its application to curation efforts in the Conserved Domain Database, Database, Volume 2019, 2019, baz064, https://doi.org/10.1093/database/baz064
Close - Share Icon Share
Abstract
This study proposes a text similarity model to help biocuration efforts of the Conserved Domain Database (CDD). CDD is a curated resource that catalogs annotated multiple sequence alignment models for ancient domains and full-length proteins. These models allow for fast searching and quick identification of conserved motifs in protein sequences via Reverse PSI-BLAST. In addition, CDD curators prepare summaries detailing the function of these conserved domains and specific protein families, based on published peer-reviewed articles. To facilitate information access for database users, it is desirable to specifically identify the referenced articles that support the assertions of curator-composed sentences. Moreover, CDD curators desire an alert system that scans the newly published literature and proposes related articles of relevance to the existing CDD records. Our approach to address these needs is a text similarity method that automatically maps a curator-written statement to candidate sentences extracted from the list of referenced articles, as well as the articles in the PubMed Central database. To evaluate this proposal, we paired CDD description sentences with the top 10 matching sentences from the literature, which were given to curators for review. Through this exercise, we discovered that we were able to map the articles in the reference list to the CDD description statements with an accuracy of 77%. In the dataset that was reviewed by curators, we were able to successfully provide references for 86% of the curator statements. In addition, we suggested new articles for curator review, which were accepted by curators to be added into the reference list at an acceptance rate of 50%. Through this process, we developed a substantial corpus of similar sentences from biomedical articles on protein sequence, structure and function research, which constitute the CDD text similarity corpus. This corpus contains 5159 sentence pairs judged for their similarity on a scale from 1 (low) to 5 (high) doubly annotated by four CDD curators. Curator-assigned similarity scores have a Pearson correlation coefficient of 0.70 and an inter-annotator agreement of 85%. To date, this is the largest biomedical text similarity resource that has been manually judged, evaluated and made publicly available to the community to foster research and development of text similarity algorithms.
Introduction
Text mining has been established as a necessary tool to help improve knowledge reusability through improved data access, representation and curation (1, 2). Biological knowledge bases rely heavily on expert curation and scaling up to accommodate the growth of the scientific literature has been a continued challenge (3). Automatically annotating biological entities such as genes/proteins and diseases and other important information in the biomedical literature, such as the dataset used in a study or the dataset location, is useful for improving the scalability of biocuration services (4, 5). The 2012 report resulting from the survey conducted for the BioCreative 2012 Workshop (Washington, DC) indicated that more databases have adopted text mining into their curation workflows compared to 2009 and seen a significant increase in curation efficiency (6).
With this study we propose to use text mining for biocuration in a capacity that has not been used before. The Conserved Domain Database (CDD; https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) is a curated resource that consists of a collection of annotated multiple sequence alignment models for ancient domains and full-length proteins (7). CDD content includes domain models curated by the CDD professional curators at NCBI, as well as models imported from external source databases. CDD-curated models use 3D-structure information to explicitly define domain boundaries and provide insights into sequence, structure and function relationships. These manually curated records are integrated within the NCBI’s search and retrieval system and are cross-linked with other databases such as Gene, 3D-structure, PubMed and PubChem. However, human curated CDD record summaries do not contain specific article references for each curated sentence, but rather a curated reference list is attached to each record summary as a whole.
The CDD curation needs are two-fold: (i) to provide an improved accessibility of the information summarized by curators for the specific conserved domains, based on their review of scientific literature, by providing reference PubMed articles for the curated sentences and (ii) to discover other (and more recently) published articles that could be brought to curators attention to review for improvement and expansion of the information presented.
To address these needs, we formulate the problem as a text similarity retrieval problem, and we describe our study with these specific contributions:
1) A method that maps the references attached to a given CDD record summary to the correct sentences related to them in the summary, for better information access;
2) A method that discovers new relevant PubMed articles for a given CDD record summary for curator review; and
3) A set of sentence pairs manually judged for similarity that can be used as a benchmark in the development of improved text similarity algorithms.
For a given query sentence, we detect semantically related sentences in PubMed. Specifically, we used this method to map the CDD record summary sentences as queries to the PubMed articles listed as references by finding the most related similar sentences within the articles. We used the same method to connect the CDD summary sentences to the best matching articles in the whole PubMed Central database.
In this manuscript, we first give an overview of the CDD database of curated protein domains, and we discuss text similarity research, its applications and related work. Next, we describe the text similarity method, the CDD text similarity corpus and give full annotation guidelines. The corpus is composed of 5159 pairs of sentences manually judged for similarity and has a high inter-annotator agreement. Finally, we show how this work was useful for CDD curation needs.
We predict that the CDD text similarity corpus, freely available to the community, will be a very important dataset for further training and testing of text similarity methods in the biomedical domain.
Background and Related Work
The CDD
CDD at NCBI is a professionally annotated resource that catalogs multiple sequence alignment models for proteins, which are available as position-specific score matrices to allow for fast identification of conserved domains in protein sequences via Reverse PSI-BLAST. The current live CDD version, v3.16, contains 56 066 protein models and protein domain models, with content obtained from Pfam (8), SMART (9), the COGs collection (10), TIGRFAMS (11), the NCBI Protein Clusters collection (12) and NCBI’s in-house data curation effort (7). Currently, CDD annotates ∼460 million or close to 78% of the sequences in NCBI’s Protein database, including 96% of structure-derived protein sequences that are over 30 residues long.
CDD curators are highly trained domain-expert professionals. They annotate functional sites, which can be mapped onto protein (query) sequences. Currently, a total of 29 991 site annotations have been created on 10 605 out of 12 805 NCBI-curated domain models. Conserved sequence patterns have been recorded for 2123 of these site annotations, and their mapping onto query sequences is contingent on pattern matches.
NCBI-curated domain models also contain summaries detailing the function of specific conserved domains as well as descriptions of specific protein families. These are based on published articles from peer-reviewed journals and these PubMed IDs are included in a list of reference articles for each domain description and also as evidence for site annotation. CDD does not embed references in the summaries. However, other users of the resource, such as InterPro (http://www.ebi.ac.uk/interpro/) that incorporates CDD data already uses this format of embedded references for their records. For this purpose, we are exploring the use of text mining as an opportunity to automatically link the references to the sentences they support to facilitate information access.
Text mining research and text similarity
Text mining research is concerned with how to design a computer algorithm to extract useful information from natural language text. As stated, the problem is very general; it becomes more tractable with a defined information need and a defined set of natural language texts as source. Problems have varied from labeling different classes of entities that appear in text (13–15) or types of relations between entities that may appear in text (16, 17), to answering specific questions posed by a user from the text (18, 19), to summarizing the text (20), to deciding if the text might contain useful information regarding a particular topic (21, 22), to deciding how similar two passages of text are (23, 24).
Our general interest is in finding sentences closely related to a sentence provided by a user as a query to a large repository of sentences. One approach to this problem would be to ask for sentences that have the same meaning as the query text. This is known as semantic textual similarity (25, 26). We have examined semantic textual similarity measures and find them too restrictive for our problem (27). We argue that any closely related text is likely to be relevant to the query text. At first glance this would appear to be a problem ideally suited for a textual entailment algorithm (28). However, any closely related text is likely to provide evidence either for or against a statement and in either case this evidence will be of interest. We therefore approach our problem as an information retrieval problem (29) and seek to find those sentences most closely related or about the focus of the query text. The details of our approach are given in the methods section below.
Methods
Text similarity method
As described in the Introduction, we approach the sentence retrieval problem as an information retrieval problem. Since sentences are short, there is reason to be concerned about the vocabulary mismatch problem, i.e. words in the query sentences may not appear in a highly relevant answer sentence. This problem can be a significant concern for short query texts as described in (24, 30, 31). However, preliminary testing suggested that exact lexical term matching and the use of term weights, i.e. a form of the vector retrieval model, was sufficient for our purposes. This approach is also much more efficient to apply to the repository of over half a billion sentences from which our answer sentences can come.
The alpha weights
Here |${c}_s$| is the number of documents in |${P}_t$| that contain |$s$| and |${w}_s$| is the Bayesian weight determined by the naïve Bayesian learning for |$s$|. Conveniently |${c}_s$| is a byproduct of the learning algorithm and entails no extra computation. Because |${c}_s/\Big\Vert {P}_t\Big\Vert$| is the fraction of documents in |${P}_t$| that contain the term |$s$|, the alpha weight is just the average Bayesian score over all documents in |${P}_t$| where the scoring is restricted to those positive-weighted terms that co-occur significantly with |$t$|. Here requiring the |$P$|-value for co-occurrence with |$t$| to be less then |$1/\Big\Vert {T}_p\Big\Vert$| is a Bonferroni correction, but likely stronger than needed.
Why should we think the alpha weight is a useful weight for retrieval of sentences? Our examination of a good deal of data computed in this way has convinced us that the most semantically meaningful terms receive the highest alpha weights. By the method of computation these are the terms that have the strongest ties to their context. Another way to say this is to state that these are the terms that predict their context, i.e. if we find them we already know what else we are likely to find. If we then match them in retrieval we are likely to find other elements that also match that are important. Because this is somewhat independent of term frequency, it gives a different result than IDF weighting. In our limited experience we have seen alpha weighting perform well, and we believe it has advantages over IDF, but it is not a completely settled issue. To illustrate, consider the word `maybe’. This word occurs in 75 061 sentences in PubMed Central and receives an alpha weight of 6.7474. Compare this with the gene name `bax’, which appears in 255 445 sentences and receives the alpha weight of 27.8179. This seems in accord with the relative importance of the two terms, whereas, if IDF weighting were used `maybe’ would receive the higher weight. Another example where the alpha weight is helpful is the case of an important word being misspelled. Consider the word `phosphatase’, which occurs in 509 366 sentences, receives the alpha weight of 27.7916. The misspelled version `phosphatese’ occurs in 40 sentences and receives the alpha weight of 8.10017. Assuming the misspelling is a random event it seems appropriate the misspelled version should lose influence. We would not want this misspelled word to dominate retrieval and preclude a top match with a sentence containing the correct spelling. The IDF weight would rate `phosphatese’ much higher than the correct spelling. In spite of these examples the alpha weight does generally give a lower weight to more frequent terms because they tend to be less specific so less predictive of their environment. Thus, the term `p53’, which occurs in 1 090 923 sentences, receives the alpha weight of 16.7673. Even though it is very important, it is not specific enough in determining its context, i.e. it appears in a large variety of contexts.
The above applies to single token terms and we refer to the result as alpha-singles weighting. To assign a weight to a term, which is a pair of tokens, we average the alpha weights of its two tokens and multiply this average by 0.2 by analogy with IDF-pairs weighting. This allows pairs of tokens to have an influence when they match, but this influence is limited in such a way that a sentence match with a higher number of single tokens matching usually receives the higher score. The pairs determine the relative ranking when the same single terms match, but one sentence also matches on pairs and receives the higher score. At least that is the intent of scoring pair matches. We believe it is a little better at matching phraseology. We refer to the resulting weighting as the alpha-pairs weighting.
Alpha-pairs weighting was used to retrieve the data for the work reported in this paper. This was done based on a previous experiment comparing alpha-pairs and IDF-pairs retrieval: a set of 1000 PMC titles was randomly chosen from the past couple of years of entries in PMC, and the titles were used as queries to pull out the top 10 sentences in the remainder of the PMC documents (the PMC document contributing the title was not allowed to contribute to the retrieved set). This retrieval was done with both IDF-pairs and alpha-pairs weighting. For each of the 1000 queries, the retrieved sets of 10 sentences each were compared, and if they differed the first rank at which they differed provided a pair of answer sentences that could be compared as answers to see which was the better answer to the query. For all but two of the query sentences there was at least one difference in the retrieved sets by the two methods. These 998 pairs were presented in a random (blinded) manner to one of the authors who is a physician, and he judged each pair on a scale of 1–5 for relevance to the query title, based on Annotation Guidelines in Appendix A. In 560 of these pairs the answer sentences received different ratings. In 312 where ratings differed, the sentence retrieved by the alpha method received the better rating. This means alpha’s performance was better than that of IDF in 56% of the cases where pair ratings differed (95% CI, 52–59%). It was on the basis of this result that the alpha method was used to create the retrieved sets studied in this paper and forming the basis for Table 3. The judgments, which are the basis for Table 3, however, were by different judges, and this clearly shows the need for more study of the alpha method.
Normalized discounted cumulative gain
Because we are using a multipoint relevance scale with five levels, we evaluate performance using normalized discounted cumulative gain (nDCG) (34) as our performance measure. Since our judgment scale is 1–5 and 1 means not at all relevant, we decrease all values on the scale by 1 so non-relevance corresponds to |${2}^0-1=0$| in the standard formula (https://en.wikipedia.org/wiki/Discounted_cumulative_gain).
Cumulative Gain (CG) is defined as |${CG}_p=\sum_{i=1}^p({2}^{r_i}-1)$|, at a particular rank position p, where ri is the graded relevance of the result at position i. CG does not include the position of a result in the consideration of the usefulness of a result set. As such, the value computed with the CG function is unaffected by changes in the ordering of search results. That is, moving a highly relevant document above a higher ranked, less relevant, document does not change the computed value for CG.
Discounted Cumulative Gain (DCG), defined as |${DCG}_p=\sum_{i=1}^p\frac{2^{r_i}-1}{{\mathit{\log}}_2(i+1)}$|, at a particular rank position p, accounts for the usefulness of the search results by placing stronger emphasis on retrieving relevant documents, and penalizing highly relevant documents appearing lower in a search result list as the graded relevance value is reduced logarithmically proportional to the position of the result.
Because search results vary depending on the query, comparing a search engine’s performance from one query to the next cannot be consistently achieved using DCGp alone, so the cumulative gain for a chosen value of p should be normalized across queries. This is done by sorting all relevant documents in the corpus by their relative relevance, producing the maximum possible DCGp through position p, also called Ideal DCGp (IDCGp) through that position. For a query, the normalized discounted cumulative gain, or nDCGp, is computed as follows: |${nDCG}_p=\frac{DCG_p}{IDCG_p}$|.
The nDCGp values for all queries can be averaged to obtain a measure of the average performance of a search engine’s ranking algorithm. Note that in a perfect ranking algorithm, the DCGp will be the same as the ideal DCGp producing an nDCGp of 1.0. All nDCGp calculations are then relative values on the interval 0.0–1.0 and so they are cross-query comparable.
Results
Building the corpus
Here, we describe how we created the text similarity corpus with sentence pairs from the CDD record summaries and PubMed/PubMed Central articles. We also describe the tool designed to help curators review this type of data and enter their judgments. Lastly, we describe results of the annotation process and their use.
The CDD records data
In May 2018, we retrieved 12 774 conserved domain description summaries written by NCBI curators since 1999. These summaries range from 1 to 43 sentences and consist of 7.2 sentences on average. The described conserved domains have a list of PubMed references that support curator assertions. These range from 1 to 375, with an average of 11 PubMed references per CDD record. We randomly selected a set of 40 CDD records for manual annotation containing an average of 7 sentences per record and 10.7 PubMed references per record. Of these, we completed the double annotation for 37 records, which constitute the CDD sentence similarity corpus.
We followed this approach: we selected a random set of CDD records, their curator-written summaries and their lists of referenced articles. The set of referenced articles is a mix of PubMed documents and PubMed Central articles. For those PubMed articles where we were able to access the full text (32%), we used the full text; for the rest we used their PubMed title and abstract. The curator-written summaries of the CDD records and the text of the referenced articles were segmented into sentences. Then, for each sentence in the CDD summary, we calculated its similarity to every sentence in the set of referenced articles, using the text similarity method. For each query sentence, we selected up to the top 10 scoring similar sentences for manual review (but only sentences with a positive score).
Next, to discover articles of possible interest to curators for the selected CDD records, we used the whole PubMed Central database as our Reference Candidate set. We repeated the same process of calculating sentence similarity scores between the CDD summary sentences and all the sentences in the Reference Candidate set, excluding the articles that were already curated in the references list. Again, for each sentence in the given CDD summary we picked up to 10 most similar sentences for manual review.
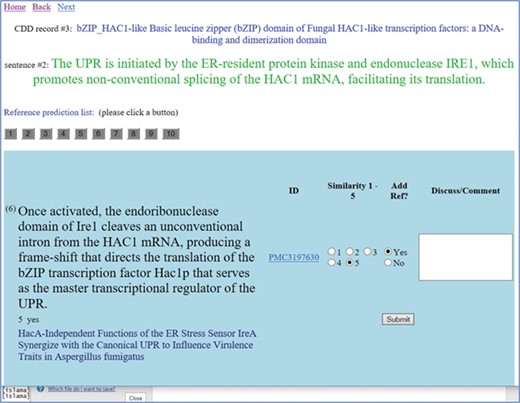
A screenshot of the text similarity annotation tool. The curator is reviewing the CDD record titled `Basic leucine zipper (bZIP) domain of Fungal HAC1-like transcription factors: a DNA-binding and dimerization domain’. The curator is reviewing the second sentence from the original summary, which is shown in green letters. Clicking on the button numbered 1–10, the reviewer can see the candidate sentences extracted from the reference list of articles. Each of these sentences is judged on a similarity scale 1–5, with 5 meaning most similar and 1 meaning the least similar. A link to the article is provided (in this case, a link to the full text is provided). The title of the referenced article is given under the candidate sentence, for the curator’s convenience. The curator selects the `Add reference’ button as needed.
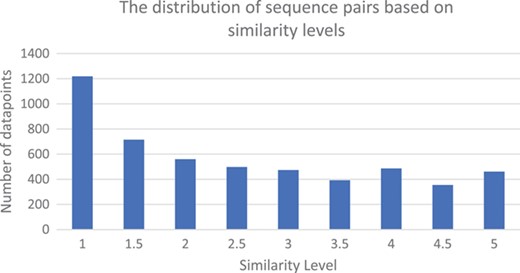
The similarity levels distribution of the judged pairs of sentences in the CDD sentence similarity dataset.
Inter-annotator agreement on Reference prediction set
| REFERENCE prediction set . | Exact agreement . | Relaxed agreement . | Agreement on reference . | |
|---|---|---|---|---|
| Curator 1 | Curator 2 | 55.18 | 86.82 | 84.18 |
| Curator 1 | Curator 3 | 46.00 | 86.00 | 85.14 |
| Curator 1 | Curator 4 | 42.81 | 87.50 | 76.56 |
| Curator 2 | Curator 3 | 54.81 | 89.26 | 83.89 |
| Curator 2 | Curator 4 | 34.15 | 79.95 | 72.09 |
| Curator 3 | Curator 4 | 32.86 | 76.12 | 65.48 |
| Average | 45.62 | 84.56 | 78.49 | |
| REFERENCE prediction set . | Exact agreement . | Relaxed agreement . | Agreement on reference . | |
|---|---|---|---|---|
| Curator 1 | Curator 2 | 55.18 | 86.82 | 84.18 |
| Curator 1 | Curator 3 | 46.00 | 86.00 | 85.14 |
| Curator 1 | Curator 4 | 42.81 | 87.50 | 76.56 |
| Curator 2 | Curator 3 | 54.81 | 89.26 | 83.89 |
| Curator 2 | Curator 4 | 34.15 | 79.95 | 72.09 |
| Curator 3 | Curator 4 | 32.86 | 76.12 | 65.48 |
| Average | 45.62 | 84.56 | 78.49 | |
Inter-annotator agreement on Reference prediction set
| REFERENCE prediction set . | Exact agreement . | Relaxed agreement . | Agreement on reference . | |
|---|---|---|---|---|
| Curator 1 | Curator 2 | 55.18 | 86.82 | 84.18 |
| Curator 1 | Curator 3 | 46.00 | 86.00 | 85.14 |
| Curator 1 | Curator 4 | 42.81 | 87.50 | 76.56 |
| Curator 2 | Curator 3 | 54.81 | 89.26 | 83.89 |
| Curator 2 | Curator 4 | 34.15 | 79.95 | 72.09 |
| Curator 3 | Curator 4 | 32.86 | 76.12 | 65.48 |
| Average | 45.62 | 84.56 | 78.49 | |
| REFERENCE prediction set . | Exact agreement . | Relaxed agreement . | Agreement on reference . | |
|---|---|---|---|---|
| Curator 1 | Curator 2 | 55.18 | 86.82 | 84.18 |
| Curator 1 | Curator 3 | 46.00 | 86.00 | 85.14 |
| Curator 1 | Curator 4 | 42.81 | 87.50 | 76.56 |
| Curator 2 | Curator 3 | 54.81 | 89.26 | 83.89 |
| Curator 2 | Curator 4 | 34.15 | 79.95 | 72.09 |
| Curator 3 | Curator 4 | 32.86 | 76.12 | 65.48 |
| Average | 45.62 | 84.56 | 78.49 | |
The annotation process and corpus development
The annotation tool is designed to provide flexibility and ease of annotation. Curators have individual login pages, and the task entry page is designed to list CDD record titles in groups of 10. Clicking on a title takes the curator to a second page that lists the CDD record description summary segmented into sentences. Then, the curator can click on each individual CDD record sentence as a query to see the selection of candidate retrieved sentences. The candidate sentences are organized in two groups. The candidate sentences selected from the set of articles referenced in the record are shown in the first block, called the `Reference prediction list’, and the candidate sentences selected from the whole set of PubMed Central articles are shown in the second block at the bottom of the page, called the `Discovery prediction list’.
We decided that a visual interface focusing on the pair of sentences being evaluated for similarity level was more convenient than a visual interface showing all extracted candidate sentences at once. The tool implemented visual clues to denote when the curator has already made a judgment. The button allowing the perusal of candidate sentences changes color if a judgment is previously recorded. The CDD record summary sentence changes color if a judgment is recorded for all candidate sentences, and the CDD record title changes color if a judgment is made for all statements in its description summary.
Curators provided two types of judgments: a similarity value ranging from 1 (low) to 5 (high), which we describe in the Annotation Guidelines, Appendix A, and a click indicating whether the article that the candidate sentence was extracted from should be linked as reference to the current summary sentence.
Four CDD curators participated in this study, and two curators manually evaluated the degree of similarity of each candidate reference sentence to the original summary sentence. At the same time, they judged whether the article containing the candidate sentence should be listed as a reference for that summary sentence. In Figure 1, we show a screenshot of the annotation tool that facilitated this manual review.
As a result of the manual curation, we have the largest dataset of similar sentences in the biomedical literature. This corpus is composed of 5159 pairs of sentences judged for similarity by two curators. These sentences originated from 37 CDD record summary descriptions, which in turn were tokenized into 259 statement sentences. Then these sentences were paired with 2571 sentences retrieved from the articles listed as references, and with 2588 sentences retrieved from the whole set of PubMed Central articles. Since each pair of sentences received two judgments, we use the average of the two judgments for evaluation purposes. The distribution of the similarity values in the CDD sentence similarity dataset is shown in Figure 2. This figure shows that this dataset is very appropriate for training and testing of automatic tools to detect text similarity, as all similarity levels are well represented.
Analysis of the annotation process
Inter-annotator agreement
To evaluate the quality of the dataset, we calculated the Pearson correlation coefficient for the reference prediction set and for the Discovery prediction set and found the values 0.678 and 0.775, respectively. These are very high Pearson correlation values and confirm the fact that the duplicate curator judgments are indeed similar and well correlated.
We also calculated the inter-annotator agreement on the exact agreement, relaxed agreement similarity level and agreement on the selection of references. The exact agreement similarity level requires both curators to pick the same similarity level for any given pair of sentences. The relaxed agreement similarity measure allows a difference of 1 in the similarity level, for example, one curator could judge a pair of sentences as a 5, while the other judges it as a 4. The agreement on the selection of references measures the frequency that both curators judge that the article the reference sentence is selected from should be listed as a reference for the summary sentence. These numbers are shown in Tables 1 and 2.
Inter-annotator agreement for Discovery prediction set
| DISCOVERY prediction set . | Exact agreement . | Relaxed agreement . | Agreement on reference . | |
|---|---|---|---|---|
| Curator 1 | Curator 2 | 62.28 | 91.00 | 86.33 |
| Curator 1 | Curator 3 | 57.27 | 86.92 | 85.76 |
| Curator 1 | Curator 4 | 49.20 | 81.03 | 80.71 |
| Curator 2 | Curator 3 | 51.58 | 85.34 | 81.45 |
| Curator 2 | Curator 4 | 58.10 | 91.52 | 87.92 |
| Curator 3 | Curator 4 | 45.43 | 81.97 | 79.39 |
| Average | 54.40 | 86.67 | 83.66 | |
| DISCOVERY prediction set . | Exact agreement . | Relaxed agreement . | Agreement on reference . | |
|---|---|---|---|---|
| Curator 1 | Curator 2 | 62.28 | 91.00 | 86.33 |
| Curator 1 | Curator 3 | 57.27 | 86.92 | 85.76 |
| Curator 1 | Curator 4 | 49.20 | 81.03 | 80.71 |
| Curator 2 | Curator 3 | 51.58 | 85.34 | 81.45 |
| Curator 2 | Curator 4 | 58.10 | 91.52 | 87.92 |
| Curator 3 | Curator 4 | 45.43 | 81.97 | 79.39 |
| Average | 54.40 | 86.67 | 83.66 | |
Inter-annotator agreement for Discovery prediction set
| DISCOVERY prediction set . | Exact agreement . | Relaxed agreement . | Agreement on reference . | |
|---|---|---|---|---|
| Curator 1 | Curator 2 | 62.28 | 91.00 | 86.33 |
| Curator 1 | Curator 3 | 57.27 | 86.92 | 85.76 |
| Curator 1 | Curator 4 | 49.20 | 81.03 | 80.71 |
| Curator 2 | Curator 3 | 51.58 | 85.34 | 81.45 |
| Curator 2 | Curator 4 | 58.10 | 91.52 | 87.92 |
| Curator 3 | Curator 4 | 45.43 | 81.97 | 79.39 |
| Average | 54.40 | 86.67 | 83.66 | |
| DISCOVERY prediction set . | Exact agreement . | Relaxed agreement . | Agreement on reference . | |
|---|---|---|---|---|
| Curator 1 | Curator 2 | 62.28 | 91.00 | 86.33 |
| Curator 1 | Curator 3 | 57.27 | 86.92 | 85.76 |
| Curator 1 | Curator 4 | 49.20 | 81.03 | 80.71 |
| Curator 2 | Curator 3 | 51.58 | 85.34 | 81.45 |
| Curator 2 | Curator 4 | 58.10 | 91.52 | 87.92 |
| Curator 3 | Curator 4 | 45.43 | 81.97 | 79.39 |
| Average | 54.40 | 86.67 | 83.66 | |
nDCG performance of the four methods on the two sets of humanly judged retrieved sentences
| . | Alpha-pairs . | Alpha-singles . | IDF-pairs . | IDF-singles . |
|---|---|---|---|---|
| Discovery prediction set | 0.7058 | 0.6740 | 0.7337 | 0.7132 |
| Reference Prediction set | 0.7413 | 0.7419 | 0.7496 | 0.7599 |
| . | Alpha-pairs . | Alpha-singles . | IDF-pairs . | IDF-singles . |
|---|---|---|---|---|
| Discovery prediction set | 0.7058 | 0.6740 | 0.7337 | 0.7132 |
| Reference Prediction set | 0.7413 | 0.7419 | 0.7496 | 0.7599 |
nDCG performance of the four methods on the two sets of humanly judged retrieved sentences
| . | Alpha-pairs . | Alpha-singles . | IDF-pairs . | IDF-singles . |
|---|---|---|---|---|
| Discovery prediction set | 0.7058 | 0.6740 | 0.7337 | 0.7132 |
| Reference Prediction set | 0.7413 | 0.7419 | 0.7496 | 0.7599 |
| . | Alpha-pairs . | Alpha-singles . | IDF-pairs . | IDF-singles . |
|---|---|---|---|---|
| Discovery prediction set | 0.7058 | 0.6740 | 0.7337 | 0.7132 |
| Reference Prediction set | 0.7413 | 0.7419 | 0.7496 | 0.7599 |
Both the Pearson correlation coefficient values, listed above, and the inter-annotator agreement measures, shown in Tables 1 and 2, show that the curators had a high degree of agreement. This agreement is reflected both in the similarity judgments that they assigned to all sentence pairs in the dataset and in the selection of references for the sentences in the CDD summary. The high agreement between the curators shows that: (i) the CDD text similarity corpus is a high-quality corpus of biomedical text similarity; and (ii) the future work for population of reference articles in the CDD record summaries could be streamlined in such a way that only one curator reviews the output of the automated text similarity method.
Usefulness of the sentence similarity method for the CDD curation
We wanted to quantify how useful this study was for the CDD curators. The first goal was to build a method that could help populate the curator-written summaries with the referenced articles listed at the end of the description. This analysis is included in full in the supplementary files, where we list the details for all the manually curated CDD records used in this study.
We measured the usefulness of text mining in matching the list of referenced articles to the sentences in the CDD record description. To evaluate this, we first counted the number of articles listed in the reference section, then we reviewed the number of articles from which the text mining method had extracted candidate sentences. Next, we counted the number of reference articles that were accepted as supporting the corresponding record sentences. In summary, from a total of 395 articles listed as references in 37 CDD records, the text mining method retrieved suggestions from 312, of which 240 were accepted by curators, for an acceptance rate of 76.92% (Supplementary file, Table S1).
Next, we counted the number of sentences in the CDD record description and reviewed the number of sentences for which at least a reference article was accepted after the manual review. Text mining had successfully provided at least one reference suggestion for all individual summary sentences in the CDD records, and of those, curators accepted at least one suggested reference for 86.54% of all summary sentences. These results are also very encouraging for the prospect of large-scale semi-automatic curation.
The second goal was to explore the discovery of new articles to bring to the curators’ attention for review and possible inclusion in the curated CDD records. A detailed view of this analysis is also included in the Supplementary file, Table S2. We reviewed the discovered articles, and we compared their publication dates with the publication date of the most recent article in the list of the record’s referenced articles. This analysis revealed that 65.6% of the suggested articles are more recent than the ones in the referenced list. The acceptance rate of curators for the discovery articles is 50%.
We reviewed some candidate sentences from both the Reference and Discovery sets, which even though they were assigned a high similarity score, were not accepted as references, and we discovered that these sentences were extracted from the related work sections and were referring to studies and results described in other articles. The future implementation could eliminate candidate sentences retrieved from related work or background sections, though in some cases these may lead a curator to useful material in other articles that would not otherwise be found.
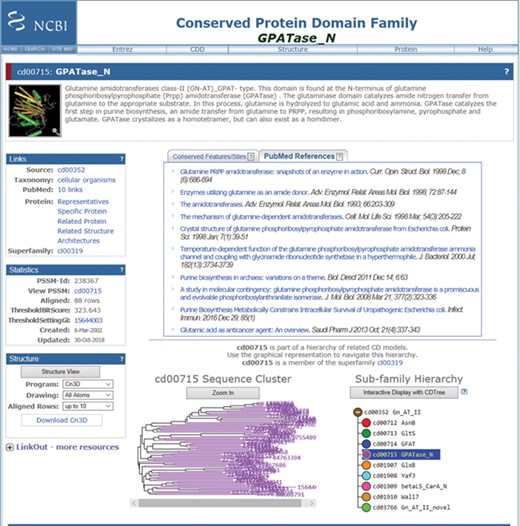
Screenshot of a random CDD record (the GPATase_N domain) page, after being updated with the new information in the CDD. This record now has 10 PubMed articles linked as relevant references. The six new references are added below the original references (https://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=cd00715).
Use of CDD text similarity corpus to benchmark different text similarity methods
The top 10 related sentences were retrieved by using the alpha-pairs method. This was done in two ways. Each query sentence was a part of a paragraph written by a curator and reference articles from PubMed were listed for each such paragraph. The first search was to search the query sentence against the PubMed Central full-text article if available, but if not, to search against the PubMed text of title and abstract. The top 10 resulting sentences were retrieved but any that had zero scores were dropped. We refer to this as the reference prediction set. Retrieval was also done with each query sentence against all the sentences in PubMed Central, and the top 10 results were retrieved and again any zeroes were dropped. We refer to this as the Discovery prediction set. On each of these sets we compared the performance of IDF-pairs, IDF-singles, Alpha-pairs and Alpha-singles using nDCG5. The results are in Table 3. First, one notes that all methods perform better on the Reference prediction set than on the Discovery prediction set. This is not surprising because the retrieval on the list of referenced articles restricts the retrieval to sentences from documents that have already been judged by the curators as appropriate references for the statements. Second, we see that the performance is better with the IDF methods. The IDF-pairs method performs better than IDF-singles on the Discovery set indicating that phraseology is an important factor in the retrieval. The fact that IDF-singles performs better on the Reference set may be due to the limited selection of sentences to rank in this case, which may leave few options for matching phraseology. We believe these sets of human judgments will prove useful in evaluating other retrieval methods and comparing them with the IDF-pairs and IDF-singles methods.
Incorporation of results on the CDD webpage
The new articles that were suggested and accepted as references for the CDD records (from the Discovery prediction set) have been manually added to the webpages for a few example conserved domains. The GPATase_N domain, cd00715, is such an example. This record originally listed only four reference articles that had been included through manual curation efforts. Text mining uncovered six additional reference articles, five of which are more current than the most recent article originally included. Figure 3 shows this updated CDD record. The description of the CDD record now incorporates the six discovered references for this CDD record.
In the near future, these references may be added using a semi-automated approach, with some curator oversight and validation. CDD records that have only a few references will be enhanced by being populated with additional references using this system. It is possible that future conserved domain records will have shorter descriptions and have a strong, succinct title, both of which could be incorporated in the search algorithm to obtain the most relevant references that can be added in this manner. In addition, the algorithm could also be further optimized for new reference discovery. For this case, the search algorithm could review the recent articles and prioritize the articles containing new information for the conserved domain.
Conclusions and future directions
The PubMed text similarity method presented here was used to help CDD curators identify relevant reference articles matching curated sentences in CDD record summaries. It was also used to identify other relevant publications (such as more recent articles) to help CDD curators review and update the curated records. This was done via identification of candidate similar sentences extracted from the list of referenced articles, and the whole PubMed Central database. That these efforts were successful is attested by the high rate of curator acceptance of the results. An important product of these efforts is the CDD text similarity corpus of 5159 pairs of sentences, which contains similarity judgments by four curators. This doubly-annotated data is of high quality as shown by the high Pearson correlation coefficient and the inter-annotator agreement.
To use this text mining similarity method to facilitate reference matching for all the CDD records, we need to implement a procedure that uses algorithm output and curator time most efficiently. Several steps are required: (1) Use the CDD text similarity corpus to improve the text similarity method, to suggest fewer candidates at the highest confidence level; (2) Build a CDD record review interface that incorporates the input of the text similarity method and allows curators to select a given record, browse the suggestions, and choose articles to add as references; and (3) Roll-out this interface to all CDD curators, to save curation time and maintain the high standard of data curation quality.
More broadly, the development of automatic methods that can efficiently detect documents of a high degree of similarity to an original text input, has a primary utility in a large set of downstream tasks where it can be used as a system component, such as question answering, information retrieval, document clustering and text summarization. To this end, the CDD text similarity corpus is an important contribution to the community and, as far as we are aware, the largest manually judged dataset of sentence pairs in the biomedical domain.
Conflict of interest. None declared.
Database URL:ftp://ftp.ncbi.nlm.nih.gov/pub/lu/Suppl/CDD/
References
A. Appendix 1 Annotation guidelines
We will be using the following CDD record summary to illustrate the annotation guidelines. We will select a sentence from the CDD record summary and will show several candidate sentences that were paired with the record sentence, and which the curators judged for similarity.
CDD record title: Protein-interacting, N-terminal, Bro1-like domain of Saccharomyces cerevisiae Bro1 and related proteinshttps://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=185765
This level of similarity denotes that if the original statement is of interest it seems quite unlikely the paired sentence would be of any interest.
Examples
CDD record sentence (query):Bro1-like domains are boomerang-shaped, and part of the domain is a tetratricopeptide repeat (TPR)-like structure.
Candidate sentence 1: Coi12p has two FK506-binding domains (FKBDs) and a tetratricopeptide repeat (TPR) domain (Fig 5A and Supplementary Fig S4).
Candidate sentence 2: The predicted structure suggests the presence of four structural domains: a tetratricopeptide repeat (TPR) domain, a beta-propeller domain, a carboxypeptidase regulatory domain-like fold (CRD), and an OmpA-C-like putative peptidoglycan-binding domain.
Notes
Both these candidate sentences have a very low similarity with the statement sentence extracted from the above CDD record. The corresponding articles as well, were not selected as references.
This level of similarity denotes that the relationship between the pair of sentences is not close, but there is some possibility of a useful relationship between the two, though there are more differences than similarities.
Examples
CDD record sentence (query):Snf7 binds to a conserved hydrophobic patch on the middle of the concave side of the Bro1 domain.
Candidate sentence 1: YPXnL-based late domains provide an elegant mechanism to support the release of viral buds because the Bro1 domain of ALIX directly binds to the SNF7 subunit of ESCRT-III.
Candidate sentence 2: Given the endosomal localization of Bro1 domain proteins Alix and Rim20, inferences from previous deletion analyses of Alix and Rim20, and the conservation of most residues in the surface identified as the Bro1 binding site for Snf7, it is reasonable to infer that the Bro1 domain is a conserved ESCRT-III targeting domain.
Notes
Both these candidate sentences have been judged to have a similarity level of 2 with the statement sentence extracted from the above CDD record. The corresponding articles also, were not selected as references.
This level of similarity denotes that there seems to be a relationship of interest between the pair of sentences, but it is uncertain if this is a useful relationship.
Examples
CDD record sentence (query):Snf7 binds to a conserved hydrophobic patch on the middle of the concave side of the Bro1 domain.
Candidate sentence 1: As a positive control, we used Snf7, which binds directly to the ‘Bro1 domain’ of Bro1 (Figure 4A).
Candidate sentence 2: The C-terminus of Snf7 contains a motif that binds to the Bro1 domain of Bro1.
Notes
Both these candidate sentences have been judged to have a similarity level of 3 with the statement sentence extracted from the above CDD record. The corresponding articles were not selected as references.
This level similarity denotes that the candidate sentences differ in some important aspects from the original statements, but they still look quite likely to be of interest in relation to the originals.
Examples
CDD record sentence (query):Snf7 binds to a conserved hydrophobic patch on the middle of the concave side of the Bro1 domain.
Candidate sentence 1: A hydrophobic patch surrounding a conserved Ile on the Bro1 domain contacts exposed hydrophobic residues from the C-terminal helix of Snf7 (Fig. 11).
Candidate sentence 2: Thus, we conclude that the association of Bro1 with Snf7, which is mediated by hydrophobic patch 1 in the Bro1 domain, is important for the localization of Bro1 to endosomal membranes.
Notes
Both these candidate sentences have been judged to have a similarity level of 4 with the statement sentence extracted from the above CDD record. The corresponding articles were both selected as references.
This level of similarity denotes that all essential elements of the original statements are contained in the candidate sentences and even though the candidates may go beyond the original in detail, the candidates are clearly useful.
Examples
CDD record sentence (query):Snf7 binds to a conserved hydrophobic patch on the middle of the concave side of the Bro1 domain.
Candidate sentence 1: We have found that Snf7 interacts with hydrophobic patch 1 on the middle of the filled-in concave side of the Bro1 domain.
Candidate sentence 2: The Bro1 domain is shaped like a banana, and binds to the C-terminal region of Snf7 through a conserved patch near the center of its concave face.
Notes
Both these candidate sentences have been judged to have a similarity level of 5 with the statement sentence extracted from the above CDD record. The corresponding articles were both selected as references.