-
PDF
- Split View
-
Views
-
Cite
Cite
Cristina Dorador, Irma Vila, Francisco Remonsellez, Johannes F. Imhoff, Karl-Paul Witzel, Unique clusters of Archaea in Salar de Huasco, an athalassohaline evaporitic basin of the Chilean Altiplano, FEMS Microbiology Ecology, Volume 73, Issue 2, August 2010, Pages 291–302, https://doi.org/10.1111/j.1574-6941.2010.00891.x
Close - Share Icon Share
Abstract
Analyses of clone libraries from water and sediments of different sites from Salar de Huasco, a high-altitude athalassohaline wetland in the Chilean Altiplano, revealed the presence of five unique clusters of uncultured Archaea that have not been previously reported or specifically assigned. These sequences were distantly related (83–96% sequence identity) to a limited number of other clone sequences and revealed no identity to cultured Archaea. The abundance of Archaea and Bacteria was estimated using qPCR and community composition was examined through the construction of clone libraries of archaeal 16S rRNA gene. Archaea were found to be dominant over Bacteria in sediments from two saline sites (sites H4: 6.31 × 104 and site H6: 1.37 × 104 μS cm−1) and in one of the water samples (freshwater from site H0: 607 μS cm−1). Euryarchaeotal sequences were more abundant than crenarchaeotal sequences. Many of the clone sequences (52%) were similar to uncultured archaeal groups found in marine ecosystems having identity values between 99% and 97%. A major fraction of the sequences (40%) were members of Methanobacteria, while others were included in the Marine Benthic Groups B and D, the Miscellaneous Crenarchaeotic Group, the Terrestrial Miscellaneous Euryarchaeotal Group, Marine Group I and Halobacteria. The presence of uncultured archaeal groups in Salar de Huasco extends their known distribution in inland waters, providing new clues about their possible function in the environment.
Introduction
Archaea are widely distributed in both extreme (e.g. hot springs, hydrothermal vents, solfataras, salt lakes, soda lakes, sewage digesters, rumen) and nonextreme (e.g. ocean waters, lakes, soil) environments (Chaban et al., 2006). The domain Archaea consists of two major phyla: Crenarchaeota and Euryarchaeota. With the advent of molecular techniques, an immense number of 16S rRNA gene sequences of ‘uncultured Archaea’ have been retrieved in clone libraries from different environments (Schleper et al., 2005). Some of these uncultured archaeal groups are known from marine environments (DeLong, 1998; Vetriani et al., 1999; Takai et al., 2001; Schleper et al., 2005; Teske & Sørensen, 2008) and represent quantitatively important members of the pelagic deep-ocean picoplankton (Karner et al., 2001). Among the Euryarchaeota, Marine Group II (marine plankton, anaerobic digestor), Marine Group III (marine sediments, marine plankton) (DeLong, 1998), Marine Benthic Group D (MBGD, deep sea and salt marsh sediments) (Vetriani et al., 1999) and the South Africa gold mine euryarchaeotic group [SAGME-1, SAGME-2, sequences also included in the Terrestrial Miscellaneous Euryarchaeotal Group (TMEG); Takai et al., 2001] have been reported frequently from terrestrial and marine environments (e.g. Inagaki et al., 2003; Shao et al., 2004; Sørensen et al., 2005; Sørensen & Teske, 2006; Kendall et al., 2007). Among the Crenarchaeota, the Marine Group I (MG-I), Marine Benthic Group A, B and Marine Benthic Group C (MBGC, deep sea sediments) were first reported from seawater (DeLong, 1992; Fuhrman et al., 1992; Vetriani et al., 1999), and more recently also from subsurface marine sediments (Sørensen et al., 2004; Teske, 2006). The Miscellaneous Crenarchaeotic Group (MCG) have a wider habitat range, which includes terrestrial and marine, hot and cold, surface and subsurface environments (Teske, 2006). Functional gene surveys and pure culture studies have indicated that at least some members of MG-I are aerobic and autotrophic ammonia oxidizers (Francis et al., 2005; Könneke et al., 2005). Studies using reverse transcription have demonstrated that several of these uncultured marine archaeal groups are metabolically active in deep subsurface sediments (Biddle et al., 2006). Intensive studies of uncultured Archaea have been conducted especially in marine environments, including metagenomic analyses (e.g. Martin-Cuadrado et al., 2008). However, little is known about their function in the environment. Various authors have described the occurrence and abundance of various archaeal groups in inland waters (e.g. Sørensen et al., 2005; Galand et al., 2006; Briée et al., 2007; Auguet & Casamayor, 2008; Jiang et al., 2008).
Salar de Huasco is located in the Chilean Altiplano at an altitude of 3800 m and exhibits salinity conditions from freshwater to saturated salt waters (Dorador et al., 2008a); it is considered an athalassohaline system because its salt composition is markedly different from that of seawater (e.g. Oren, 2006). Abiotic conditions in the Altiplano, including low temperatures (mean annual temperature <5 °C), low atmospheric pressure (40% lower than that at sea level), high solar radiation (<1100 W m−2), strong variation in different environmental properties at daily, annual and interannual time scales and a negative water balance, shape the biological communities in these water bodies (Vila & Mühlhauser, 1987). Considering the environmental conditions of this inland aquatic system, we expected to encounter a highly adapted archaeal community, including taxa adapted to high salt concentrations. In the present study, we describe the composition and abundance of archaeal assemblages in water and sediment samples at, in terms of salt concentration, four contrasting sites from Salar de Huasco using denaturing gradient gel electrophoresis (DGGE), clone libraries of the 16S rRNA gene and quantitative PCR (qPCR). In addition, the presence of ammonia-oxidizing Crenarchaeota was examined by cloning the ammonia monooxygenase gene amoA, reflecting the possible role of Archaea in the biogeochemical nitrogen cycle.
Materials and methods
Site description and sampling
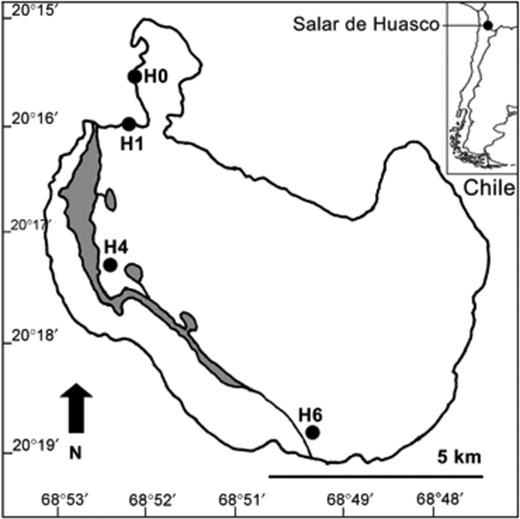
During January 2005 (summer), two samples were collected per site at four different sites from the Salar de Huasco (20°18′S, 68°50′W) (Fig. 1). Water samples were collected in plastic sterile bottles of 1 L from the surface and sediment samples were taken with a polycarbonate hand corer (30 cm length and 3 cm diameter). The surface area of the Salar extends to c. 50 km2, with open water representing only 2.5 km2, and the water level can vary seasonally (Risacher et al., 2003). The ponds are shallow and located along a N–S aspect through the Salar, forming a natural salinity gradient with freshwater sites H0 (stream) and H1 (shallow lagoon) and the saline site H4 (shallow lagoon). Site H6 (shallow lagoon) exhibited lower salinity than site H4 because the water comes from a different freshwater stream (Risacher et al., 1999). A summary of some key morphometric, physical and chemical characteristics of the sampling sites is presented in Table 1. A more detailed description of their properties was described in Dorador. (2008a).
Map indicating the location of Salar de Huasco and four study sites (H0, H1, H4 and H6). Gray areas indicate the presence of permanent lagoons.
Physical and chemical characteristics of water samples and microbial abundance at the four sites in Salar de Huasco
| Characteristics | Sites | |||
| H0 | H1 | H4 | H6 | |
| Location | 20°15′32″, 68°52′25″ | 20°16′08″, 68°52′29″ | 20°17′41″, 68°53′00″ | 20°19′43″, 68°50′19″ |
| Altitude (m) | 3799 | 3795 | 3789 | 3789 |
| Type | Stream | Lagoon | Lagoon | Lagoon |
| Conductivity (μS cm−1) | 607 | 645 | 6.31 × 104 | 1.37 × 104 |
| Total dissolved salts (g L−1) | 0.42 | 0.46 | 64.93 | 9.38 |
| Dissolved oxygen (mg L−1) | 6.9 | 10.3 | 0 | 8.4 |
| pH | 7.7 | 8.7 | 8.2 | 8.8 |
| Temperature (°C) | 16.6 | 19 | 20.1 | 12 |
| Hour | 14:30 | 16:00 | 15:50 | 10:15 |
| N-NO3− (μg L−1) | 55.5 | 53.5 | 60 | 30 |
| P-PO43− (μg L−1) | 40.3 | 20.3 | 3.91 × 103 | 807 |
| S-SO42− (mg L−1) | 22.1 | 26.6 | 3990 | 141.2 |
| Si (mg L−1) | 19.9 | 12.4 | 25.4 | 18.1 |
| Hardness (mg L−1) | 162.5 | 130 | 3500 | 1000 |
| Total alkalinity (mM) | 2 | 1.52 | 12 | 7 |
| Chla (μg L−1) | 2.2 | 8.5 | 39 | 114.3 |
| Cations | Ca2+>Na+>K+>Mg2+ | Ca2+>Na+>K+>Mg2+ | Mg2+>Ca2+>K+>Na+ | Ca2+>Mg2+>Na+>K+ |
| Archaeal abundance water (cells mL−1) | 69 | 174 | 1.68 × 104 | 7.8 × 104 |
| Archaeal abundance sediment (cells g−1) | 3.24 × 106 | 5.04 × 107 | 3.6 × 107 | 5.94 × 106 |
| Bacterial abundance water (cells mL−1) | 45 | 288 | 5.34 × 105 | 2.7 × 106 |
| Bacterial abundance sediment (cells g−1) | 2.16 × 108 | 5.94 × 108 | 1.08 × 107 | 2.52 × 106 |
| Characteristics | Sites | |||
| H0 | H1 | H4 | H6 | |
| Location | 20°15′32″, 68°52′25″ | 20°16′08″, 68°52′29″ | 20°17′41″, 68°53′00″ | 20°19′43″, 68°50′19″ |
| Altitude (m) | 3799 | 3795 | 3789 | 3789 |
| Type | Stream | Lagoon | Lagoon | Lagoon |
| Conductivity (μS cm−1) | 607 | 645 | 6.31 × 104 | 1.37 × 104 |
| Total dissolved salts (g L−1) | 0.42 | 0.46 | 64.93 | 9.38 |
| Dissolved oxygen (mg L−1) | 6.9 | 10.3 | 0 | 8.4 |
| pH | 7.7 | 8.7 | 8.2 | 8.8 |
| Temperature (°C) | 16.6 | 19 | 20.1 | 12 |
| Hour | 14:30 | 16:00 | 15:50 | 10:15 |
| N-NO3− (μg L−1) | 55.5 | 53.5 | 60 | 30 |
| P-PO43− (μg L−1) | 40.3 | 20.3 | 3.91 × 103 | 807 |
| S-SO42− (mg L−1) | 22.1 | 26.6 | 3990 | 141.2 |
| Si (mg L−1) | 19.9 | 12.4 | 25.4 | 18.1 |
| Hardness (mg L−1) | 162.5 | 130 | 3500 | 1000 |
| Total alkalinity (mM) | 2 | 1.52 | 12 | 7 |
| Chla (μg L−1) | 2.2 | 8.5 | 39 | 114.3 |
| Cations | Ca2+>Na+>K+>Mg2+ | Ca2+>Na+>K+>Mg2+ | Mg2+>Ca2+>K+>Na+ | Ca2+>Mg2+>Na+>K+ |
| Archaeal abundance water (cells mL−1) | 69 | 174 | 1.68 × 104 | 7.8 × 104 |
| Archaeal abundance sediment (cells g−1) | 3.24 × 106 | 5.04 × 107 | 3.6 × 107 | 5.94 × 106 |
| Bacterial abundance water (cells mL−1) | 45 | 288 | 5.34 × 105 | 2.7 × 106 |
| Bacterial abundance sediment (cells g−1) | 2.16 × 108 | 5.94 × 108 | 1.08 × 107 | 2.52 × 106 |
Physical and chemical characteristics of water samples and microbial abundance at the four sites in Salar de Huasco
| Characteristics | Sites | |||
| H0 | H1 | H4 | H6 | |
| Location | 20°15′32″, 68°52′25″ | 20°16′08″, 68°52′29″ | 20°17′41″, 68°53′00″ | 20°19′43″, 68°50′19″ |
| Altitude (m) | 3799 | 3795 | 3789 | 3789 |
| Type | Stream | Lagoon | Lagoon | Lagoon |
| Conductivity (μS cm−1) | 607 | 645 | 6.31 × 104 | 1.37 × 104 |
| Total dissolved salts (g L−1) | 0.42 | 0.46 | 64.93 | 9.38 |
| Dissolved oxygen (mg L−1) | 6.9 | 10.3 | 0 | 8.4 |
| pH | 7.7 | 8.7 | 8.2 | 8.8 |
| Temperature (°C) | 16.6 | 19 | 20.1 | 12 |
| Hour | 14:30 | 16:00 | 15:50 | 10:15 |
| N-NO3− (μg L−1) | 55.5 | 53.5 | 60 | 30 |
| P-PO43− (μg L−1) | 40.3 | 20.3 | 3.91 × 103 | 807 |
| S-SO42− (mg L−1) | 22.1 | 26.6 | 3990 | 141.2 |
| Si (mg L−1) | 19.9 | 12.4 | 25.4 | 18.1 |
| Hardness (mg L−1) | 162.5 | 130 | 3500 | 1000 |
| Total alkalinity (mM) | 2 | 1.52 | 12 | 7 |
| Chla (μg L−1) | 2.2 | 8.5 | 39 | 114.3 |
| Cations | Ca2+>Na+>K+>Mg2+ | Ca2+>Na+>K+>Mg2+ | Mg2+>Ca2+>K+>Na+ | Ca2+>Mg2+>Na+>K+ |
| Archaeal abundance water (cells mL−1) | 69 | 174 | 1.68 × 104 | 7.8 × 104 |
| Archaeal abundance sediment (cells g−1) | 3.24 × 106 | 5.04 × 107 | 3.6 × 107 | 5.94 × 106 |
| Bacterial abundance water (cells mL−1) | 45 | 288 | 5.34 × 105 | 2.7 × 106 |
| Bacterial abundance sediment (cells g−1) | 2.16 × 108 | 5.94 × 108 | 1.08 × 107 | 2.52 × 106 |
| Characteristics | Sites | |||
| H0 | H1 | H4 | H6 | |
| Location | 20°15′32″, 68°52′25″ | 20°16′08″, 68°52′29″ | 20°17′41″, 68°53′00″ | 20°19′43″, 68°50′19″ |
| Altitude (m) | 3799 | 3795 | 3789 | 3789 |
| Type | Stream | Lagoon | Lagoon | Lagoon |
| Conductivity (μS cm−1) | 607 | 645 | 6.31 × 104 | 1.37 × 104 |
| Total dissolved salts (g L−1) | 0.42 | 0.46 | 64.93 | 9.38 |
| Dissolved oxygen (mg L−1) | 6.9 | 10.3 | 0 | 8.4 |
| pH | 7.7 | 8.7 | 8.2 | 8.8 |
| Temperature (°C) | 16.6 | 19 | 20.1 | 12 |
| Hour | 14:30 | 16:00 | 15:50 | 10:15 |
| N-NO3− (μg L−1) | 55.5 | 53.5 | 60 | 30 |
| P-PO43− (μg L−1) | 40.3 | 20.3 | 3.91 × 103 | 807 |
| S-SO42− (mg L−1) | 22.1 | 26.6 | 3990 | 141.2 |
| Si (mg L−1) | 19.9 | 12.4 | 25.4 | 18.1 |
| Hardness (mg L−1) | 162.5 | 130 | 3500 | 1000 |
| Total alkalinity (mM) | 2 | 1.52 | 12 | 7 |
| Chla (μg L−1) | 2.2 | 8.5 | 39 | 114.3 |
| Cations | Ca2+>Na+>K+>Mg2+ | Ca2+>Na+>K+>Mg2+ | Mg2+>Ca2+>K+>Na+ | Ca2+>Mg2+>Na+>K+ |
| Archaeal abundance water (cells mL−1) | 69 | 174 | 1.68 × 104 | 7.8 × 104 |
| Archaeal abundance sediment (cells g−1) | 3.24 × 106 | 5.04 × 107 | 3.6 × 107 | 5.94 × 106 |
| Bacterial abundance water (cells mL−1) | 45 | 288 | 5.34 × 105 | 2.7 × 106 |
| Bacterial abundance sediment (cells g−1) | 2.16 × 108 | 5.94 × 108 | 1.08 × 107 | 2.52 × 106 |
DNA extraction and PCR amplifications
Environmental DNA was extracted from water and sediment samples from each site. For DNA extraction, water samples were filtered on-site in a 0.22-μL pore-size filter without prefiltration, with the volume of filtered water ranging between 0.05 L for saline sites (sites H4 and H6) and 1 L for the freshwater site (site H0). For DNA extractions of sediment samples, we used 600 mg of homogenized sediment. All DNA extractions were carried out using the UltraClean Soil DNA isolation kit (MoBio Laboratories Inc.) according to the manufacturer's instructions.
Amplification of 16S rRNA archaeal gene was performed using a nested PCR approach. Fragments of 1500 bp were obtained with primers Ar4F (positions 8–25) and Un1492R, and then primers Ar3F (positions 7–26)–Ar9R (positions 906–927) (Jurgens et al., 2000) were used to amplify the 16S rRNA gene from Archaea with the first-round PCR products as templates in a nested PCR. The reverse primer used, Ar9R (Jurgens et al., 2000), exhibited mismatches with the uncultured groups SAGMEG, Marine Hydrothermal Vent Group (MHVG, Takai & Horikoshi, 1999), Ancient Archaeal Group (AAG, Takai & Horikoshi, 1999) and Deep-Sea Archaeal Group (DSAG, Inagaki et al., 2003) (Teske & Sørensen, 2008), which can possibly underestimate the presence of these groups in environmental samples. Each PCR reaction contained 10 × PCR buffer with 2 mM MgCl2 (Roche), 200 μM dNTP mixture (Gibco), 1 pmol of each primer, 2.5 U Taq polymerase (Roche), 10–100 ng template DNA and water to a final volume of 50 μL. The PCR conditions used were initial denaturation at 94 °C for 5 min, 35 cycles of denaturation (30 s at 94 °C), annealing (45 s at 55 °C) and extension (1.5 min at 73 °C). Archaeal amoA gene fragments were amplified by PCR using primers CrenAmo1F and CrenAmo1R (Könneke et al., 2005) and the following PCR conditions: initial denaturation at 94 °C for 3 min; 30 cycles of denaturation (30 s at 94 °C), annealing (30 s at 56 °C) and extension (3 min at 72 °C); and a final extension at 72 °C for 1 min. For cloning, PCR reactions were carried out with Pfu polymerase (Promega) following the manufacturer's instructions.
DGGE analysis
DGGE was performed according to Casamayor et al., (2000) with PCR products of archaeal 16S rRNA gene generated in a nested approach. Primers 344F (Raskin et al., 1994) and 915R (Stahl & Amann, 1991) were used to amplify the archaeal 16S rRNA gene PCR product. The 344F primer contained an additional 40 nucleotide GC-rich sequence (GC clamp) at its 5′-end in order to maintain stable melting behavior during DGGE (Muyzer et al., 1993). The PCR conditions were as described above and PCR amplification was carried out using a touchdown protocol as follows: an initial denaturing step of 5 min at 94 °C, 20 cycles of 30 s at 94 °C, 45 s at 65–55 °C (decreased by 0.5 °C every cycle) and 1.5 min at 72 °C, and then 10 cycles of 30 s at 94 °C, 45 s at 55 °C and 1.5 min at 72 °C. PCR products were applied onto 7.5% polyacrylamide gels containing a linear gradient of 30–60% denaturant where 100% denaturant was defined as 7 M urea and 40% formamide. DGGE was carried out in the BioRad D Gene System (Bio-Rad Laboratories, Hercules, CA) at 60 °C, 200 V, for 6 h. Gels were stained with the SYBR Gold nucleic acid gel stain (Molecular Probes). In order to examine the relationships between communities in the different samples, a matrix was constructed from the distribution pattern of the bands in different samples, and cluster analyses (UPGMA), based on percent similarity between the samples, were conducted using the multivariate statistical package (msvp, version 3.12d; Kovach Computing Services, Wales, UK).
Cloning and 16S rRNA gene sequence analysis
Archaeal clone libraries were generated from water and sediment samples collected from the four study sites. Samples for cloning (one sample per site) were selected according to their richness detected by DGGE (data not shown). Purified amplicons were cloned into pCR-Blunt vector (Invitrogen) according to the manufacturer's instructions. Analysis of the inserts, sequencing and sequence analysis were described previously (Dorador et al., 2008b). Briefly, sequences were checked for chimeras, rarefaction curves were constructed and nonparametric richness estimators SACE and SChao1 and the Shannon–Weaver diversity index were determined manually and via the web interface available at http://www.aslo.org/lomethods/free/2004/0114a.html (Kemp & Aller, 2004).
Phylogenetic analysis
The phylogenetic affiliations of the archaeal sequences obtained here were estimated as described elsewhere (Dorador et al., 2008b). Sequences were aligned using the alignment tool of the arb package (http://www.arb-home.de), a maximum likelihood analysis in the program phyml (Guindon et al., 2005) with the GTR substitution model and 100 bootstrap resamplings. Trees were edited using mega4 (Tamura et al., 2007). Sequences with similarities >97% were considered to represent the same phylotype (Stackebrandt & Goebel, 1994).
Nucleotide sequence accession numbers
The nucleotide sequences from this study are available in GenBank (http://www.ncbi.nlm.nih.gov) under accession numbers EU481526–EU481630 for 16S rRNA gene sequences and FJ839431–FJ839434 for archaeal amoA sequences.
qPCR
qPCR was conducted using SYBR-Green PCR Master Mix (Biotools) in an MJ Mini-Opticon thermocycler (Bio-Rad). Primers for Archaea (ARCH349F and ARCH806R) and Bacteria (UBactF and UBactR) (Takai & Horikoshi, 2000; Nadkarni et al., 2002) were used for the quantification of total archaeal and bacterial 16S rRNA genes, respectively. The reaction mixture contained 10 μL of SYBR-Green PCR Master Mix (Biotools), 1 μL of template DNA (∼10 ng), 1 μL of the corresponding oligonucleotide primers (final concentration of 0.25 μM) and nuclease-free H2O added to a total of 20 μL. The Archaea amplification program consisted of an initial denaturing at 95 °C for 10 min, and then 40 cycles of 95 °C for 35 s, 56 °C for 40 s and 72 °C for 20 s. The Bacteria amplification program consisted of an initial denaturing at 95 °C for 10 min, and then 40 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 20 s. For both amplification programs, the fluorescence measurements were recorded at the end of each extension step and the melting curves were measured with a ramp increasing the temperature from 50 to 95 °C by 1 °C every 5 s. All analyses were performed in triplicate including a negative control.
Standard curves and data analysis
To generate bacterial and archaeal standard curves, we extracted plasmid DNA from 16S rRNA gene clones containing Escherichia coli and clone Hua1-w87, respectively. The plasmid DNA was quantified using gel electrophoresis with a DNA low-mass ladder (Promega). Standard curves for bacterial and archaeal 16S rRNA genes were constructed using serial dilutions of these plasmids' DNA, ranging from 850 to 0.0085 pg, as a template. The DNA concentrations in ng μL−1 were transformed to 16S rRNA gene copy number μL−1, by means of the following transformation: first, we obtained the molecular weight of DNA (plasmid+insert), assuming that 660 is the average molecular weight of a base pair. We divided Avogadro's number (6.02 × 1023) by this molecular weight to estimate the abundance of molecules per 1 g. Finally, this value was divided by 1 × 109 to obtain the gene copy number per ng of DNA. The calibration curves were generated using the opticon monitor™ ver. 3.1.32 software (Bio-Rad Laboratories), and for each standard, the concentration was plotted against the cycle number, and the value at which the fluorescence signal increased above the threshold value was the cycle threshold (Ct value). The bacterial and archaeal standard curve showed an efficiency of 1.23 (R2 value of 0.995) and 0.91 (R2 value of 0.999), respectively.
Results
Microbial abundance in Salar de Huasco and quantification of 16S rRNA genes
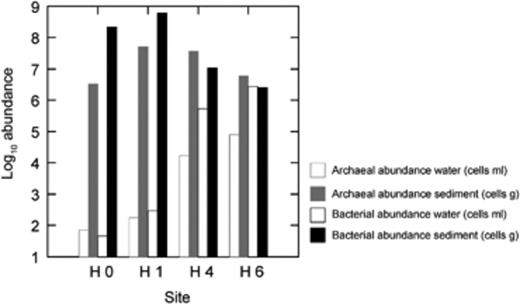
The total numbers of microbial cells in water and sediment samples of sites H0 and H1 were calculated by qPCR results assuming one copy of 16S rRNA gene per cell and were 101 and 102 cells mL−1 and 106–108 cells g−1, respectively. For sites H4 and H6, these values fluctuated between 104 and 106 cells mL−1 for water and 106 and 107 cells g−1 for sediment (Table 1). The estimated abundance of Bacteria and Archaea in waters and sediments from the four sites of Salar de Huasco is shown in Fig. 2. There was a clear differentiation between water and sediment samples throughout the salinity gradient from site H0 (freshwater stream) to H4 (saline lagoon) (Fig. 2).
Relative abundance of Bacteria and Archaea in Salar de Huasco at the sampling sites, determined by qPCR.
Construction of 16S rRNA gene clone libraries and estimation of archaeal richness
Clone libraries of 16S rRNA genes were constructed from sites H0, H1, H4 and H6. In total, 137 clones from water samples and 197 from sediments were obtained. Rarefaction curves from the four sites showed saturation at low phylotype numbers, from 4 to 11 in water samples and from 10 to 32 phylotypes in sediment samples (Supporting Information, Fig. S1). The richness estimators SACE and SChao1 (Chao, 1984; Chao, 1987) provided estimates of the total number of phylotypes ranging from 5 to 44 (SACE) and 4 to 26 (SChao1) in water samples and from 23 to 107 and 18 to 91 in sediment samples (Table 2). The Shannon–Weaver diversity index indicated a higher archaeal diversity in sediments (1.4–3.0) compared with the water samples (0.7–1.7). The number of DGGE bands ranged between 4 and 16 in water samples and between 9 and 10 in sediment samples, which are lower than the richness estimated by clone libraries, and did not show differences between water and sediment samples. Nevertheless, the composition of the DGGE bands shows one cluster that largely consists of sediment samples but also includes the water sample from site H6 (Fig. S2). The pattern of bands from water samples collected from different sites within the Salar de Huasco showed no clear grouping, indicating that they were only distantly related. For example, water samples from H4 and H1 were <60%, and the water sample from H0 was <50% similar to water samples from all the other sites.
Number of clones and phylotypes, richness estimators SACE and SChao1, number of bands in DGGE and Shannon diversity (H′) in the libraries of 16S rRNA gene from water and sediment
| Site | Clone library | Number of clones in library | Number of phylotypes observed | Predicted value of SACE | Predicted value of SChao1 | Shannon diversity index (H′) | Number of bands in DGGE |
| H0 | Water | 27 | 4 | 5 | 4 | 0.7 | 16 |
| H1 | Water | 56 | 6 | 8 | 7 | 1.1 | 4 |
| H4 | Water | 33 | 11 | 44 | 26 | 1.6 | 10 |
| H6 | Water | 21 | 8 | 17 | 10 | 1.7 | 7 |
| H0 | Sediment | 64 | 23 | 34 | 30 | 2.8 | 10 |
| H1 | Sediment | 35 | 13 | 23 | 18 | 2.1 | 10 |
| H4 | Sediment | 45 | 10 | 36 | 32 | 1.4 | 9 |
| H6 | Sediment | 53 | 32 | 107 | 91 | 3 | 9 |
| Site | Clone library | Number of clones in library | Number of phylotypes observed | Predicted value of SACE | Predicted value of SChao1 | Shannon diversity index (H′) | Number of bands in DGGE |
| H0 | Water | 27 | 4 | 5 | 4 | 0.7 | 16 |
| H1 | Water | 56 | 6 | 8 | 7 | 1.1 | 4 |
| H4 | Water | 33 | 11 | 44 | 26 | 1.6 | 10 |
| H6 | Water | 21 | 8 | 17 | 10 | 1.7 | 7 |
| H0 | Sediment | 64 | 23 | 34 | 30 | 2.8 | 10 |
| H1 | Sediment | 35 | 13 | 23 | 18 | 2.1 | 10 |
| H4 | Sediment | 45 | 10 | 36 | 32 | 1.4 | 9 |
| H6 | Sediment | 53 | 32 | 107 | 91 | 3 | 9 |
Number of clones and phylotypes, richness estimators SACE and SChao1, number of bands in DGGE and Shannon diversity (H′) in the libraries of 16S rRNA gene from water and sediment
| Site | Clone library | Number of clones in library | Number of phylotypes observed | Predicted value of SACE | Predicted value of SChao1 | Shannon diversity index (H′) | Number of bands in DGGE |
| H0 | Water | 27 | 4 | 5 | 4 | 0.7 | 16 |
| H1 | Water | 56 | 6 | 8 | 7 | 1.1 | 4 |
| H4 | Water | 33 | 11 | 44 | 26 | 1.6 | 10 |
| H6 | Water | 21 | 8 | 17 | 10 | 1.7 | 7 |
| H0 | Sediment | 64 | 23 | 34 | 30 | 2.8 | 10 |
| H1 | Sediment | 35 | 13 | 23 | 18 | 2.1 | 10 |
| H4 | Sediment | 45 | 10 | 36 | 32 | 1.4 | 9 |
| H6 | Sediment | 53 | 32 | 107 | 91 | 3 | 9 |
| Site | Clone library | Number of clones in library | Number of phylotypes observed | Predicted value of SACE | Predicted value of SChao1 | Shannon diversity index (H′) | Number of bands in DGGE |
| H0 | Water | 27 | 4 | 5 | 4 | 0.7 | 16 |
| H1 | Water | 56 | 6 | 8 | 7 | 1.1 | 4 |
| H4 | Water | 33 | 11 | 44 | 26 | 1.6 | 10 |
| H6 | Water | 21 | 8 | 17 | 10 | 1.7 | 7 |
| H0 | Sediment | 64 | 23 | 34 | 30 | 2.8 | 10 |
| H1 | Sediment | 35 | 13 | 23 | 18 | 2.1 | 10 |
| H4 | Sediment | 45 | 10 | 36 | 32 | 1.4 | 9 |
| H6 | Sediment | 53 | 32 | 107 | 91 | 3 | 9 |
Phylogenetic analysis of archaeal communities in water and sediment
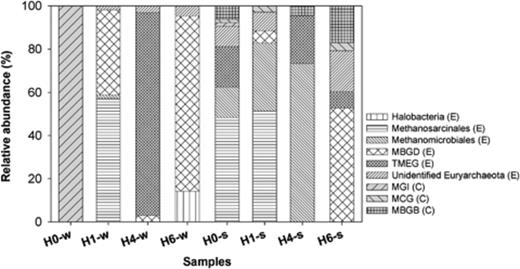
Crenarchaeota were absent from the water samples (the exception was site H0 with clones exclusively belonging to MG-I) and represented a minor fraction in the sediments. Euryarchaeota dominated in the sediments and were the exclusive archaeal representatives in water, except site H0, although different groups dominated in the different water samples. All sediments had a clearly different composition as compared with the corresponding water samples (Fig. 3 and Fig. S4).
Composition of clone libraries of 16S rRNA gene from water and sediment samples. Affiliation of the phylogenetic groups is indicated for Euryarchaeota (E) and Crenarchaeota (C). Group designations: MBGD, Marine Benthic Group D; MBGB, Marine Benthic Group B (Vetriani et al., 1999); TMEG, Terrestrial Miscellaneous Euryarchaeotal Group (Takai et al., 2001); MG-I, Marine Group I (DeLong, 1998); MCG, Miscellaneous Crenarchaeotic Group (Teske, 2006).
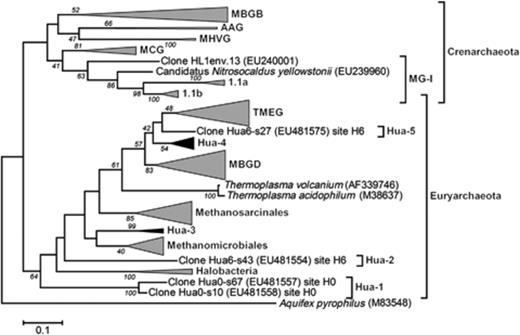
Crenarchaeota identified by clone sequences represented 14% of all clones and belonged to different phylogenetic groups. All clones from H0-w (four phylotypes) and a single phylotype from H0-s were affiliated to MG-I (Fig. S4) and clustered in Group I.1b and Group I.1a (DeLong, 1998; Schleper et al., 2005), respectively. This cluster includes sequences from soils, sediments, freshwater and subsurface sediments. Sequences from H0-w were 97% similar to an uncultured crenarchaeon retrieved from a radioactive thermal spring in the Austrian Alps (Weidler et al., 2007) and with the moderately thermophilic crenarchaeote Candidatus Nitrososphaera gargensis (Hatzenpichler et al., 2008). Representatives of the MCG were found in sediments from sites H0, H1 and H6 (four phylotypes). Clone QLW1200-A33 from Qinghai Lake (Jiang et al., 2008) exhibited 97% sequence identity with the phylotypes Hua6-s39 and Hua6-s29. Also, the MBGB was found only in sediment samples (H0-s, H4-s, H6-s). Databases contain sequences of MGBG retrieved from subseafloor sediments, lake sediments and hydrothermal vents (Fig. 4 and Fig. S4).
Phylogenetic tree based on partial 16S rRNA gene sequences (∼800 bp) of phylotypes of Archaea in water calculated by maximum likelihood analysis. Group designations: MG-I, Marine Group I (DeLong, 1998); MCG, Miscellaneous Crenarchaeotic Group (Teske, 2006); MHVG, Marine Hydrothermal Vent Group (Takai & Horikoshi, 1999); AAG, Ancient Archaeal Group (Takai & Horikoshi, 1999); MBGB, Marine Benthic Group B (Vetriani et al., 1999); MBGD, Marine Benthic Group D; TMEG, Terrestrial Miscellaneous Euryarchaeotal Group (Takai et al., 2001); Hua-1, Hua-2, Hua-3, Hua-4, Hua-5, Unidentified Euryarchaeota clusters. The scale bar represents a 10% nucleotide sequence difference. Aquifex pyrophilus (M83548) was used as an outgroup. Groups in gray include sequences from Salar de Huasco. Groups in black largely consist of sequences from Salar de Huasco. Figure S4 shows the detailed tree.
Most of the clone sequences from water and sediment samples were affiliated to various groups of Euryarchaeota (Figs 3 and 4): Halobacteria, Methanosarcinales, Methanomicrobiales, MBGD, TMEG and a group of unidentified Euryarchaeota (7% of the clones). Methanogenic Archaea were most frequently detected in Salar de Huasco (40% of the clones) (Fig. 3).
Halobacteria were represented by a single phylotype Hua6-w15, the closest cultured relative (89% sequence identity) of which was Halorubrum lipolyticum (Cui et al., 2006) isolated from Aibi salt lake in Xin-Jiang, China.
Methanosarcinales represented 25% of all clones and were found in samples H1-w, H0-s and H1-s (Fig. 3 and Fig. S4). The most frequent clone sequence from H1 (phylotype Hua1-s2) was 97% similar to Methanosaeta concilii (Eggen et al., 1989), while the most abundant clone from H0 (phylotype Hua0-s82) was 98% similar to Methanomethylovorans hollandica. This methanogen was isolated previously from freshwater sediment and utilizes dimethyl sulfide as a carbon and energy source (Lomans et al., 1999). Clone Hua0-s95 was 94% similar to Methanolobus oregonensis, isolated from anoxic, subsurface sediments of a saline, alkaline aquifer near Alkali Lake in Oregon, USA (Liu et al., 1990).
Methanomicrobiales were represented by 16% of the clones. Members of this group were found in sediment samples from the sites H0, H1 and H4, but a single clone (Hua1-w46) was also retrieved from the water of site H1 (Fig. S4). Clone Hua4-s2 was the most abundant clone from the sediment sample of site H4 and exhibited 95% sequence identity to the clone MH1492_4F retrieved from a minerotrophic fen (Cadillo-Quiroz et al., 2008). This clone also had 94% sequence identity to the Candidatus Methanoregula boonei, an acidiphilic methanogen isolated from an acidic peat bog (Bräuer et al., 2006). Clone Hua1-s38 exhibited 96% sequence identity to Methanolinea tarda (Sakai et al., 2007), a methanogen isolated from rice paddy fields that is a member of the Rice Cluster I.
The MBGD was represented by 21% of all clones and was found in sites H1, H4 and H6 (Fig. 3 and Fig. S4), with most clone sequences obtained from site H6 (81% of the clones from water and 53% from the sediment). Sequences of this cluster were reported previously from subsurface sediments, marine sediments, water and sediments of Qinghai Lake and various other habitats. The abundant clones Hua1-w90 and Hua6-w21 exhibited 91% and 97% sequence identity to the clone BCMS-5 described from prawn farm sediments in China (Shao et al., 2004).
The TMEG was first established by clones from South African gold mines (SAGMA; Takai et al., 2001), but also included sequences retrieved from subsurface and marine sediments. Sequences of TMEG comprised 17% of all clones. This group was clearly predominant in the water of site H4, but was absent from other water samples (Fig. 3). Clone Hua4-w4 had 89% sequence identity to the clone ArcA08 previously retrieved from a hypersaline endoevaporitic microbial mat in Israel and was reported as a member of the ‘Halophilic Cluster 2’ (Sørensen et al., 2005). In the sediments, TMEG was absent from H1, but present in the other sites, representing 11 phylotypes, with Hua0-s78 being closely related (97% sequence identity) to the clone WCHD3-02, previously found in a hydrocarbon and chlorinated solvent-contaminated aquifer (Dojka et al., 1998).
Unidentified Euryarchaeota that formed novel, and to date, unreported groups, which did not cluster with previously described groups, for example archaeal DGGE sequences previously reported from other wetlands of northern Chile (Demergasso et al., 2004) and that represented 7% of all clones, were found in six out of eight samples (Fig. 3). They formed five different groups (Fig. 4 and Fig. S4). Group Hua-1 consisted of two phylotypes from sediment samples of site H0 exhibiting a low identity (83–84%) to their closest relative, the clone SBAK-mid-46 retrieved from marine sediments of the Skan Bay in Alaska, classified as ‘other Euryarchaeota’ (Kendall et al., 2007). Group Hua-2 was represented by a single phylotype Hua6-s43, which exhibited 83% sequence identity to a clone of the same library (SBAK-shallow-04) classified as ‘unaffiliated Euryarchaeota’ (Kendall et al., 2007). Group Hua-3 was formed by phylotypes Hua1-w46 and Hua1-s26 together with three sequences retrieved previously from wastewater sludge, lake sediments and an anaerobic reactor (Fig. S4). Group Hua-4 is represented by the largest number of unidentified sequences and was found in H4-w, H6-w, H0-s, H1-s and H6-s. Additional sequences included in this group were recorded from marine sediments, from a sulfur spring and from petroleum-contaminated soil. The two clones from marine sediments (Skan Bay, Alaska) were similar to two different phylotypes of this study. The phylotype sequence Hua6-s37 was 96% similar to the clone SBAK-shallow-06 classified as MBGD (Kendall et al., 2007). Group Hua-5 included a single phylotype (Hua6-s27) that had 90% sequence identity to clone MKCSB-C12, which was the closest relative and was retrieved from mangrove soil (Yan et al., 2006).
Evidence of ammonia-oxidizing Archaea (AOA) in Salar de Huasco
To evaluate a possible role of Archaea in the nitrogen cycle of Salar de Huasco, we tested for the presence of AOA in all samples. PCR products of archaeal amoA, the gene coding for ammonia monooxygenase subunit A, were only obtained from the water of site H0. A clone library produced from these products consisted of four phylotypes that were affiliated to sequences retrieved from sediment and water samples (Table 3). Three phylotypes exhibited high identity (97–99%) to two clones of a clone library constructed from water samples from a drinking water distribution system (Van der Wielen et al., 2009). The fourth phylotype Hua0-w51 was 99% similar to the clone QLS1-399-A8 retrieved from sediment samples of Qinghai Lake in China (Jiang, 2009).
| Clone | Type | First hit in blast | Similarity with NM (%) | ||
| Similarity (%) | Clone name | Habitat | |||
| Hua0-w20 | Nucleotide | 97 | Clone PLANTB AR RSF-II OTU3 (EU852699) | Drinking water distribution system | 89 |
| Hua0-w51 | Nucleotide | 99 | Clone QLS1-399-A8 (EU197155) | Sediment, Qinghai Lake, Tibet, China | 74 |
| Hua0-w79 | Nucleotide | 97 | Clone PLANTB AR RSF-II OTU3 (EU852699) | Drinking water distribution system | 89 |
| Hua0-w92 | Nucleotide | 99 | Clone PLANTB AR DN OTU5 (EU852705) | Drinking water distribution system | 90 |
| Hua0-w20 | Protein | 99 | Clone PLANTA AR DN OTU4 (ACF71541) | Drinking water distribution system | 94 |
| Hua0-w51 | Protein | 100 | Clone QLS1-399-A6 (ABZ01900) | Sediment, Qinghai Lake, Tibet, China | 82 |
| Hua0-w79 | Protein | 100 | Clone PLANTA AR DN OTU4 (ACF71541) | Drinking water distribution system | 94 |
| Hua0-w92 | Protein | 100 | Clone PLANTB AR DN OTU5 (ACF71557) | Drinking water distribution system | 94 |
| Clone | Type | First hit in blast | Similarity with NM (%) | ||
| Similarity (%) | Clone name | Habitat | |||
| Hua0-w20 | Nucleotide | 97 | Clone PLANTB AR RSF-II OTU3 (EU852699) | Drinking water distribution system | 89 |
| Hua0-w51 | Nucleotide | 99 | Clone QLS1-399-A8 (EU197155) | Sediment, Qinghai Lake, Tibet, China | 74 |
| Hua0-w79 | Nucleotide | 97 | Clone PLANTB AR RSF-II OTU3 (EU852699) | Drinking water distribution system | 89 |
| Hua0-w92 | Nucleotide | 99 | Clone PLANTB AR DN OTU5 (EU852705) | Drinking water distribution system | 90 |
| Hua0-w20 | Protein | 99 | Clone PLANTA AR DN OTU4 (ACF71541) | Drinking water distribution system | 94 |
| Hua0-w51 | Protein | 100 | Clone QLS1-399-A6 (ABZ01900) | Sediment, Qinghai Lake, Tibet, China | 82 |
| Hua0-w79 | Protein | 100 | Clone PLANTA AR DN OTU4 (ACF71541) | Drinking water distribution system | 94 |
| Hua0-w92 | Protein | 100 | Clone PLANTB AR DN OTU5 (ACF71557) | Drinking water distribution system | 94 |
| Clone | Type | First hit in blast | Similarity with NM (%) | ||
| Similarity (%) | Clone name | Habitat | |||
| Hua0-w20 | Nucleotide | 97 | Clone PLANTB AR RSF-II OTU3 (EU852699) | Drinking water distribution system | 89 |
| Hua0-w51 | Nucleotide | 99 | Clone QLS1-399-A8 (EU197155) | Sediment, Qinghai Lake, Tibet, China | 74 |
| Hua0-w79 | Nucleotide | 97 | Clone PLANTB AR RSF-II OTU3 (EU852699) | Drinking water distribution system | 89 |
| Hua0-w92 | Nucleotide | 99 | Clone PLANTB AR DN OTU5 (EU852705) | Drinking water distribution system | 90 |
| Hua0-w20 | Protein | 99 | Clone PLANTA AR DN OTU4 (ACF71541) | Drinking water distribution system | 94 |
| Hua0-w51 | Protein | 100 | Clone QLS1-399-A6 (ABZ01900) | Sediment, Qinghai Lake, Tibet, China | 82 |
| Hua0-w79 | Protein | 100 | Clone PLANTA AR DN OTU4 (ACF71541) | Drinking water distribution system | 94 |
| Hua0-w92 | Protein | 100 | Clone PLANTB AR DN OTU5 (ACF71557) | Drinking water distribution system | 94 |
| Clone | Type | First hit in blast | Similarity with NM (%) | ||
| Similarity (%) | Clone name | Habitat | |||
| Hua0-w20 | Nucleotide | 97 | Clone PLANTB AR RSF-II OTU3 (EU852699) | Drinking water distribution system | 89 |
| Hua0-w51 | Nucleotide | 99 | Clone QLS1-399-A8 (EU197155) | Sediment, Qinghai Lake, Tibet, China | 74 |
| Hua0-w79 | Nucleotide | 97 | Clone PLANTB AR RSF-II OTU3 (EU852699) | Drinking water distribution system | 89 |
| Hua0-w92 | Nucleotide | 99 | Clone PLANTB AR DN OTU5 (EU852705) | Drinking water distribution system | 90 |
| Hua0-w20 | Protein | 99 | Clone PLANTA AR DN OTU4 (ACF71541) | Drinking water distribution system | 94 |
| Hua0-w51 | Protein | 100 | Clone QLS1-399-A6 (ABZ01900) | Sediment, Qinghai Lake, Tibet, China | 82 |
| Hua0-w79 | Protein | 100 | Clone PLANTA AR DN OTU4 (ACF71541) | Drinking water distribution system | 94 |
| Hua0-w92 | Protein | 100 | Clone PLANTB AR DN OTU5 (ACF71557) | Drinking water distribution system | 94 |
Discussion
Novelty of the archaeal sequences
A large portion of the sequences were not closely related to any cultured archaeon, and 24% of the sequences had similarities lower than 98% to their closest relatives. Interestingly, the rest of the sequences had clear phylogenetic affiliations, including previously described clusters for uncultured Archaea. We described five groups of unidentified Euryarchaeota, which exclusively contained representatives from Salar de Huasco (Hua-1, Hua-2 and Hua-5), and of two distinct groups containing representatives from Salar de Huasco (Hua-3 and Hua-4), together with single clone sequences from other sources (Fig. 4 and Fig. S4). Important clusters of other groups (Halobacteria, Methanomicrobiales and TMEG) including representatives from Salar de Huasco were only distantly related to the sequences available in the databases.
In prokaryotic taxonomy, an identity threshold of 97% is generally accepted in order to separate species (Stackebrandt & Goebel, 1994) and this threshold has even been increased to >98% (Stackebrandt & Ebers, 2006). Consequently, the five clusters described for uncultured Euryarchaeota from Salar de Huasco exhibited similarities lower than 96% to their closest relatives, highlighting the novelty of the sequences retrieved from this location. These results justify considering Salar de Huasco as a unique archaeal habitat and a suitable location for further, more detailed studies. The novelty of sequences and clusters has been reported recently for Cyanobacteria in Salar de Huasco (Dorador et al., 2008a, b), for Bacteria in Laguna Tebenquiche at the Salar de Atacama (Demergasso et al., 2008) and for Bacteroidetes in different saline evaporitic basins of northern Chile (Dorador et al., 2009). Along with the current work, this highlights that wetlands located in northern Chile represent potential reservoirs of undescribed microorganisms, a trend probably due to the unusual physicochemical conditions and geographical isolation of the wetlands, as well as a lack of knowledge regarding the microbial diversity in these environments.
Community composition of Archaea in Salar de Huasco
Salar de Huasco could be considered as a moderately athalassohaline wetland (Table 1), comparable in its phylogenetic groups with other high-altitude cold aquatic systems (e.g. Tibetan Lakes: Dong et al., 2006; Jiang et al., 2008) or cold saline lakes (e.g. Antarctic lakes: Karr et al., 2006). The abundance and composition of archaeal communities is likely to be driven by the availability of water, and hence, dissolution or concentration effects. The dominance of Archaea over Bacteria has been reported in habitats with NaCl concentrations close to saturation, where Halobacteria are the dominant aerobic heterotrophs (Oren, 2002). However, the levels of Archaea detected in this study (summer sampling), including the sediments of the most saline sites, were not associated with halophilic Archaea, but with uncultured marine archaeal groups and methanogens. The presence of extremely halophilic Archaea would be predicted only for sites with salt concentrations higher than 150 g L−1 (Ventosa et al., 1998). Such situations can be found in particular within the dry season when elevated salt concentrations occur due to strong evaporation. Previous studies had described elevated proportions of halophilic Archaea in water samples collected from both Salar de Huasco (site H1) and Salar de Ascotán, from northern Chile during winter (dry season) (Dorador, 2007). However, only a single phylotype was reported as a member of the Halobacteria in the present study (Figs 3 and 4, and Fig. S4). As the primers and amplification conditions used were the same in both studies, this suggests that these differences reflect temporal climatic variations (e.g. variable salt concentration).
The presence of uncultured archaeal groups in Salar de Huasco is an interesting finding, because these groups were primarily described from marine environments as an important component of the deep-ocean picoplankton (DeLong, 1998). Nevertheless, recently, uncultured marine archaeal groups were also reported from Qinghai Lake, an athalassohaline lake located in the Tibetan plateau, and in other nonmarine saline environments (Sørensen et al., 2005; Yan et al., 2006; Jiang et al., 2008), extending the distribution of these groups.
Crenarchaeota
Recovered sequences related to Crenarchaeota were affiliated to marine uncultured archaeal groups with as yet unknown functions in the environment. The only mesophile cultured representative of MG-I, Nitrosopumilus maritimus (Group 1.1a), grows by aerobically oxidizing ammonia to nitrite, a metabolic activity previously unknown for Archaea (Könneke et al., 2005). Also, members of the MG-I have been reported to demonstrate autotrophic carbon fixation while some can be mixotrophic (Ingalls et al., 2006). In this context, it is interesting that we have found MG-I members in the water of site H0, where the amoA gene was also detected (Table 3), although the majority of the 16S rRnA gene sequences were not similar (<90%) to those of Nitrosopumilus sequences, currently the only mesophile representative known to oxidize ammonia. Recently, two new thermophilic members of MG-I (Group 1.1b) that can oxidize ammonia have been described. The moderately thermophilic (46 °C) Candidatus N. gargensis (Hatzenpichler et al., 2008) enriched from hot spring microbial mats exhibited a higher sequence identity (97%) than N. maritimus with Salar de Huasco sequences, while the other thermophilic (74 °C) Candidatus Nitrosocaldus yellowstonii (de la Torre et al., 2008) retrieved from hot spring sediments showed lower similarity (<90%).
Members of the MBGB and MCG have been proposed as the major oxidizers of methane without assimilation of methane-derived carbon in subsurface sediments off Peru (Biddle et al., 2006). The presence of MBGB (sediments of H0, H4 and H6) and MCG (sediments of H0, H1 and H6) confirms the cosmopolitan distribution of this group. Because of their presence under diverse geochemical conditions, it is not possible at present to correlate their presence to particular environmental conditions.
Euryarchaeota
Sequences of MBGD were particularly abundant in water and sediment samples from site H6 and also water samples from H1. This group (equivalent to Marine Group III, as defined by DeLong, 1998) has been reported as being metabolically active in subsurface sediments (e.g. Sørensen & Teske, 2006), and recently, a metagenomic analysis of bathypelagic plankton revealed that members of Marine Group III Euryarchaeota might also utilize the oxidation of ammonia to gain energy (Martin-Cuadrado et al., 2008). The TMEG group was also reported as being metabolically active in subsurface sediments (e.g. Sørensen & Teske, 2006), but a specific physiological function of this group has not yet been found.
Role of Archaea in high-altitude saline wetlands: methanogenesis and evidence of archaeal ammonia oxidizers
Methanogenic Archaea dominated the 16S rRNA gene clone libraries of sediment samples from sites H0, H1 and H4, and probably represents an indicator of elevated methanogenic activity. In sediments, the sequences clustered with four genera of Methanosarcinales: Methanosarcina, Methanosaeta (aceticlastic methanogens), Methanomethylovorans and Methanolobus (methylotrophic organisms), highlighting the diverse substrates used by methanogens in these environments. Considering the high sulfate concentrations reported in Salar de Huasco (Risacher et al., 1999), sulfate reduction could be expected, especially at the saline sites H4 and H6, where the presence of 16S rRNA gene sequences related to sulfate-reducing bacteria has been reported previously (Dorador, 2007). Site H4 was anoxic at the time of sampling (Table 1) and exhibited the highest sulfate concentration in water (4 g L−1, almost double that of seawater, for example Holmer & Storkholm, 2001); under these conditions, almost all sequences from water samples matched into the TMEG (Fig. 3). Contrastingly, in sediment samples, Methanomicrobiales were the most abundant group (77% of the sequences). These contrasting differences in the water chemistry can help in future studies to understand the functional role of uncultured Archaea and the interaction with methanogens and sulfate-reducing bacteria. Also, it is interesting to note the low identity of the methanogenic clone sequences with the described species, highlighting the potential discovery of new species of methanogenic Archaea.
The presence of methanogens (e.g. Methanosarcinales) and benthic Archaea (MBGD) in water samples from site H1, which exhibited dissolved oxygen concentrations of 10 mg L−1, is intriguing. The water levels in different sites of Salar de Huasco are shallow (few cm) and the typically strong afternoon winds in the Altiplano can result in a well-mixed water column, and therefore the presence of anaerobic and benthic microorganisms in water samples. At site H6, methanogens were absent from clone libraries, and the most abundant group in water and sediment samples was MBGD. The functional role of this group is not yet known. Recently, it has been demonstrated that microorganisms that perform anaerobic oxidation of methane (AOM) can use alternative electron acceptors to sulfate, including Mn and Fe. It has been shown experimentally that the MBGD represents the most abundant group in anaerobic sediments treated with different electron acceptors, indicating a potential role in methane cycling (Beal et al., 2009). Interestingly, site H6 receives water from a stream different from the other sites of Salar de Huasco (Risacher et al., 1999) and has contrasting water chemistry (Fig. S3), indicating that the chemical conditions of the water (Table 1) might favor the development of members of the MBGD instead of the methanogens recorded from other sites.
Nitrogen limitation has been reported in several wetlands in the Altiplano and has also been reported for the sites H1 and H4 in Salar de Huasco (Dorador et al., 2008a). The study of the N-cycle in this system has included analysis of enrichment cultures of ammonia-oxidizing bacteria (AOB) that are dominated by Nitrosomonas (Dorador et al., 2008a). Here, we reported AOA sequences from site H0 from Salar de Huasco, indicating the potential role of this archaeal group in the ammonia oxidation. AOA has also been reported as being more abundant than AOB in other saline mountain lakes (Qinghai Lake: Jiang et al., 2009). Specific studies related to the functional role of uncultured Archaea (including AOA) are promising in Salar de Huasco because of the exceptional environmental conditions.
Acknowledgements
We thank Carolina Vargas for help with sample collection. We are grateful to Conny Burghardt and Annika Busekow for technical assistance. Special thanks are due to Chris Harrod for English corrections and statistical help and two anonymous reviewers, who helped to improve the quality of the manuscript. C.D. was supported by a doctoral fellowship from the German Academic Exchange Service (DAAD).
References
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Author notes
Editor: Michael Wagner