-
PDF
- Split View
-
Views
-
Cite
Cite
Steven Y. C. Tong, Emma J. Bishop, Rachael A. Lilliebridge, Allen C. Cheng, Zornitsa Spasova-Penkova, Deborah C. Holt, Philip M. Giffard, Malcolm I. McDonald, Bart J. Currie, Craig S. Boutlis, Community-Associated Strains of Methicillin-Resistant Staphylococcus aureus and Methicillin-Susceptible S. aureus in Indigenous Northern Australia: Epidemiology and Outcomes, The Journal of Infectious Diseases, Volume 199, Issue 10, 15 May 2009, Pages 1461–1470, https://doi.org/10.1086/598218
Close - Share Icon Share
Abstract
BackgroundSome strains of non–multidrug-resistant, methicillin-resistant Staphylococcus aureus (nmMRSA) in Australia are likely to have emerged from strains of methicillin-susceptible S. aureus (MSSA) in remote Aboriginal communities
ObjectiveTo describe the clinical epidemiology of infection due to community-associated MRSA strains in an Australian tropical hospital setting with a significant Aboriginal population and to compare infections caused by community-associated strains of MRSA, health-care–associated strains of MRSA, and MSSA strains with respect to demographic risk factors and clinical outcomes
MethodsWe queried the microbiology database for the Top End of the Northern Territory, Australia, to determine population incidences for S. aureus infection and conducted a prospective matched case-control study to compare infection due to nmMRSA, MSSA, or multidrug-resistant MRSA at the Royal Darwin Hospital
ResultsThe annual incidence of S. aureus bacteremia was 65 cases per 100,000 population, but in the Aboriginal population the incidence was 172 cases per 100,000 population (odds ratio [OR] compared with non-Aboriginal population, 5.8 [95% confidence interval {CI}, 3.8–8.9). Female sex (adjusted OR [aOR], 1.5 [95% CI, 1.1–2.0) and remote residence (aOR, 1.8 [95% CI, 1.2–2.5]) were associated with the isolation of nmMRSA rather than MSSA, but disease spectrum and outcomes were similar. Among those from whom nmMRSA was isolated, Aboriginal patients were younger (aOR for each additional year, 0.94 [95% CI, 0.92–0.96]), more likely to be female (aOR, 3.8 [95% CI, 1.7–8.5]), and more likely to reside in a remote community (aOR, 29 [95% CI, 8.9–94]) than non-Aboriginal patients. The presence of Panton-Valentine leukocidin in nmMRSA was associated with double the odds of sepsis (aOR, 2.2 [95% CI, 1.1–4.6])
ConclusionsThe association of nmMRSA infection with female sex and remote residence supports the hypothesis that nmMRSA arose from MSSA strains in remote Aboriginal communities where staphylococcal disease is highly prevalent. The similar clinical spectrum and outcomes for nmMRSA infection and MSSA infection suggest that virulence is not correlated with resistance phenotype
Infection due to community-associated strains of methicillin-resistant Staphylococcus aureus (CA-MRSA strains) was first recognized in Australia in people from remote Aboriginal communities [1]. Reports of genetically distinct CA-MRSA strains from other geographically removed regions followed [2–4]. The parallel emergence of infection due to CA-MRSA strains in North America also included outbreaks in rural Indigenous communities [5, 6]. Despite the possibility that CA-MRSA strains emerged from such communities and the impact of these strains on their inhabitants, there have been no detailed studies comparing the clinical epidemiology of infection due to CA-MRSA strains in Indigenous and nonindigenous people
Early studies noted clear differences between CA-MRSA strains and hospital-associated strains of MRSA (HA-MRSA strains) with respect to the patients affected and types of infection caused. Individuals infected with CA-MRSA strains were younger and more likely to have skin and soft-tissue infections (SSTIs), and Indigenous people were overrepresented in this group [5, 6]. In contrast, there were notable similarities between infections due to CA-MRSA strains and infections due to methicillin-susceptible S. aureus (MSSA) strains with regard to risk factors and clinical illness [5, 7]. Molecular studies demonstrated that HA-MRSA and CA-MRSA strains were genetically distinct [5, 8]. However, CA-MRSA strains and MSSA strains with the same or similar genetic backgrounds cocirculated in the same environment [9, 10]. Accordingly, it has been proposed that CA-MRSA strains emerge locally from prevalent circulating MSSA strains via acquisition of the mecA gene [11–13]
We conducted a case-control study at the Royal Darwin Hospital (RDH) with the following 2 objectives: (1) to describe the clinical epidemiology of infection due to CA-MRSA strains in an Australian tropical hospital setting with a significant Aboriginal population, and (2) to compare infections caused by CA-MRSA, HA-MRSA, and MSSA strains with respect to demographic risk factors and clinical outcomes. We hypothesized that infection due to CA-MRSA strains and infection due to MSSA strains would be similar in most of these key aspects
Methods
Study setting and designThe 330-bed RDH is located in the Top End of the Northern Territory of Australia at a tropical latitude of 12°23′ S. It is the tertiary referral center for the city of Darwin, 2 regional hospitals, and over 70 remote communities, serving a population of 176,000 and an area of 510,000 km2. Aboriginal Australians comprise 27% of the regional population but make up >50% of the RDH inpatient and emergency department population. Approval for the study was granted by the Northern Territory Department of Health and Community Services Human Research Ethics Committee and the Menzies School of Health Research
We sought nonduplicate clinical isolates of S. aureus over a 12-month period from 18 April 2006 through 17 April 2007 by querying the microbiology database that links all 3 Top End hospitals. There was no routine hospital screening for MRSA colonization during the study period. We considered non–multidrug-resistant MRSA (nmMRSA) isolates to represent CA-MRSA strains and considered multidrug-resistant MRSA (mMRSA) isolates to represent HA-MRSA strains [14]. nmMRSA isolates were defined phenotypically as those resistant to <3 non–β-lactam antibiotic classes, and mMRSA isolates were defines as those resistant to ⩾3 non–β-lactam antibiotic classes. Antibiotic phenotype has previously been shown to accurately predict the genotype of CA-MRSA strains [15], and this result has been validated in recent studies from Queensland [14], Western Australia [16], and an Australia-wide survey [12] that included isolates from Darwin. After excluding nonresident patients, we calculated incidence rates for the isolation of S. aureus on the basis of published population data from 2006 [17] and correlated this with measures of regional socioeconomic disadvantage [18] and residential remoteness [19]. For the former analysis, only regions that had ⩾10 S. aureus isolates and an available score for the index of relative socioeconomic disadvantage were included
We then conducted a case-control study of patients at the RDH in which we compared nmMRSA infections with infections caused by MSSA and mMRSA over the same 12-month period. All nmMRSA and mMRSA isolates newly identified in the microbiology laboratory were collected daily, as well as ⩽4 consecutive MSSA isolates for each nmMRSA isolate. Laboratory staff did not include isolates from patients known to have had S. aureus isolated in the past month. We used sequential laboratory receipt numbers to match eligible patients who had nmMRSA isolated to the next 2 eligible patients who had MSSA isolated
Data collectionBy prospectively reviewing the clinical records of the patients from whom each of the isolates were recovered, we collected information concerning demographic characteristics, health care–associated risk factors, comorbidities, clinical details of the infection, treatment details, and outcome at discharge. To restrict our analysis to incident staphylococcal infections, we excluded patients from further analysis if they had previously been enrolled in the study or if they had a prior existing unresolved S. aureus infection. We excluded patients who had only attended an outpatient clinic or dialysis unit and did not require treatment at the emergency department or hospital admission. We also excluded patients transferred to the RDH who had been admitted elsewhere for >24 h, patients for whom the treatment intention was palliative, and patients for whom clinical notes were unavailable
Health-care associated infection was defined in accordance with previously published criteria [20] and included nosocomial infection or the presence of any of the following risk factors during the year prior to collection of the sample that yielded the culture result of interest in the present study: (1) residence in a long-term care facility, (2) prior admission to an acute care facility, (3) use of central intravenous catheters or long-term venous access devices, (4) use of urinary catheters, (5) use of other long-term percutaneous devices, (6) prior surgical procedures, and/or (7) need for any form of dialysis. Nosocomial infection was defined by an isolate obtained from a sample collected >48 h after hospital admission. Previously published criteria were used to define infection, sepsis, and the systemic inflammatory response syndrome [21]. We defined colonization as a positive microbiological culture result in the absence of clinical features of infection
Laboratory methodsThe laboratory identified S. aureus isolates by use of standard methods and conducted susceptibility testing using an automated system (Vitek 2 V4.01; bioMérieux) and the Kirby-Bauer disk diffusion method in accordance with the guidelines of the Clinical and Laboratory Standards Institute [22]. A disk-approximation test was used to detect inducible clindamycin resistance. Real-time polymerase chain reaction was used to verify the identity of nmMRSA isolates by confirming the presence of the nucA and mecA genes; it was also used to determine the presence of pvl genes [10]
Statistical analysisStatistical significance for crude analysis of dichotomous variables was determined using the χ2 test or Fisher’s exact test. Nonparametric data were compared using the Mann-Whitney U test. We performed multivariate conditional logistic regression analysis with stepwise backward elimination of variables to identify the risk factors associated with infection due to nmMRSA compared with those for infection due to MSSA, and we performed multivariate logistic regression analysis of risk factors associated with severity of illness. The likelihood ratio test was used to assess the statistical significance of candidate risk factors. We examined the association between incidence of S. aureus infection and measures of regional socioeconomic disadvantage and remoteness by linear regression. If obvious outliers were present we used robust regression methods. Two-sided P values of <.05 were considered significant. Statistical analysis was performed with Stata (version 9.2; StataCorp)
Results
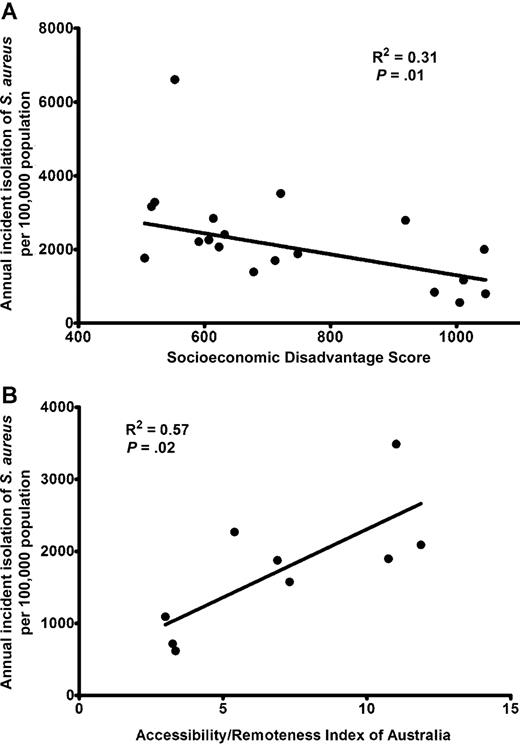
From 18 April 2006 through 17 April 2007, there were 2227 S. aureus isolates recovered from distinct patients from the 3 Top End hospitals; nmMRSA accounted for 343 (15%) of the isolates, MSSA accounted for 1748 (79%), and mMRSA accounted for 136 (6%). There were 110 (5%) nonduplicate isolates recovered from blood culture, which were distributed as follows: 17 were nmMRSA, 83 were MSSA, and 10 were mMRSA. The annual incidence of S. aureus bacteremia was 65 cases per 100,000 population, and for infection due to MRSA, the incidence was 16 cases per 100,000 population. The annual incidence of S. aureus bacteremia in the Aboriginal population was 172 cases per 100,000 population, and for the non-Aboriginal population, the incidence was 30 cases per 100,000 population (odds ratio [OR], 5.8 [95% confidence interval {CI}, 3.8–8.9]). The total annual incidence of isolation of S. aureus was 1248 isolations per 100,000 population, and the incidence of nmMRSA isolation was 193 isolations per 100,000 population. There was a strong correlation between incident isolation of S. aureus and measures of regional socioeconomic disadvantage and remoteness (figure 1)
Incident isolation of Staphylococcus aureus from 18 April 2006 through 17 April 2007 in the Top End of the Northern Territory, Australia, plotted against the regional index of relative socioeconomic disadvantage score (A) and the accessibility/remoteness index of Australia (B). For the socioeconomic score, a lower score indicates greater disadvantage; for the accessibility/remoteness index, a higher score indicates greater remoteness. Regression was performed using robust methods (A) and standard linear regression (B)
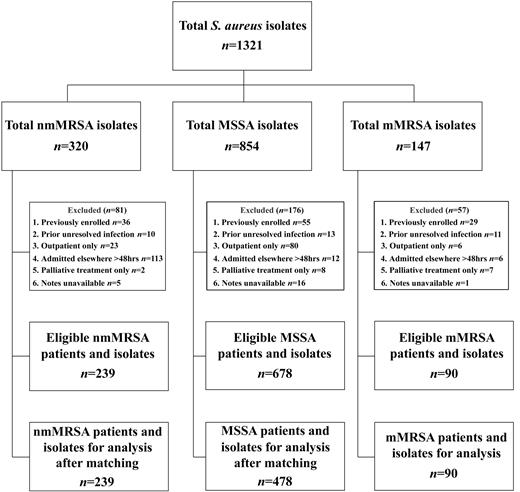
At the RDH during the same time period, there were 1693 nonduplicate S. aureus isolates recovered; 291 (17%) of these were nmMRSA, 1265 (75%) were MSSA, and 137 were (8%) mMRSA. As an estimate of community-onset cases, of 728 isolates recovered from patients presenting to the emergency department, 117 (16%) were nmMRSA, 582 (80%) were MSSA, and 29 (4%) were mMRSA. Prior to matching, there were 1007 eligible patients and isolates in the case-control study. Figure 2 presents details about exclusion criteria and the number of participants and isolates at each stage of the study. After matching, there were 239 patients who had nmMRSA isolated, 478 who had MSSA isolated, and 90 who had mMRSA isolated available for further analysis. The demographic characteristics of the control patients who had MSSA isolated were representative of the overall population of patients infected with MSSA in the Top End (data not shown). Of the 807 isolates studied, 403 (50%) were considered to represent community-associated strains, and 404 (50%) were considered to represent health care–associated strains. There were 153 (19%) isolates recovered from patients who satisfied the definition of nosocomial infection. A similar proportion of nmMRSA and MSSA isolates were health-care associated strains (P=.15) or were nosocomially acquired (P=.12) (table 1). In nmMRSA isolates, the presence of pvl genes was associated with community acquisition rather than nosocomial acquisition (OR, 4.4 [95% CI, 2.5–7.8])
Flow chart detailing the no. of Staphylococcus aureus isolates collected, the exclusion criteria applied, the no. of eligible isolates, and the no. of matched isolates used for the Royal Darwin Hospital case-control study, 18 April 2006–17 April 2007. nmMRSA, non–multidrug-resistant, methicillin-resistant Staphylococcus aureus; mMRSA, multidrug-resistant MRSA; MSSA, methicillin-susceptible S. aureus
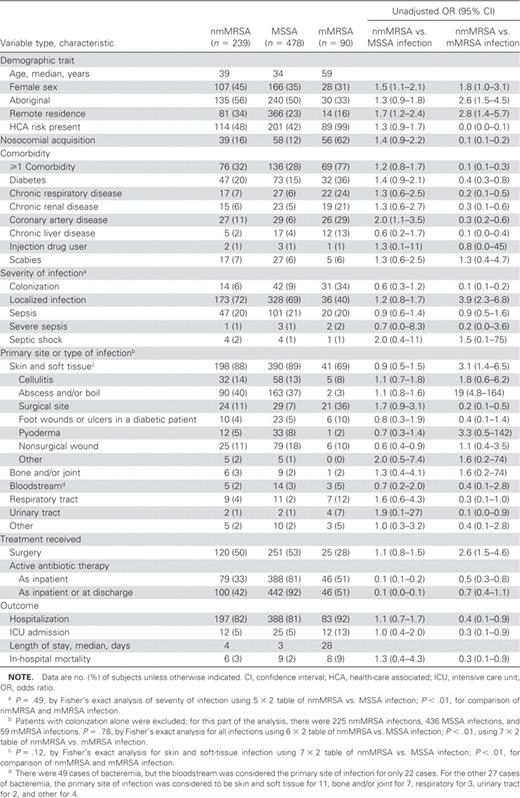
Characteristics of patients from whom non–multidrug-resistant, methicillin-resistant Staphylococcus aureus (nmMRSA), methicillin-susceptible S. aureus (MSSA), or multidrug-resistant MRSA (mMRSA) was isolated
Epidemiological and clinical characteristicsPatients who had nmMRSA isolated and patients who had MSSA isolated were of a similar age (P=.10) but younger than those who had mMRSA isolated (P<.01) (table 1). Patients who had nmMRSA isolated were less likely to be male than those who had MSSA or mMRSA isolated. A similar proportion of patients who had nmMRSA isolated and patients who had MSSA isolated were of Aboriginal ethnicity, but patients who had nmMRSA isolated were more likely to reside remotely. Otherwise, similarities between patients who had nmMRSA isolated and those who had MSSA isolated were evident with regard to the presence of comorbidities and laboratory indices. The patients who had mMRSA isolated were clearly a distinct group, being more likely to be hospitalized with comorbidities as well as having significantly higher serum creatinine levels and lower albumin and hematocrit levels (data not shown)
Severity and primary site of infectionThe distribution of clinical syndromes and severity of illness is shown in table 1. The vast majority of infections were SSTIs. Overall 629 (87%) of 720 infections (87 colonized patients were excluded) were SSTIs, accounting for ∼90% of nmMRSA and MSSA infections (198 of 225 infections and 390 of 436 infections, respectively) and 41 (69%) of 59 mMRSA infections. Boils, abscesses, and nonsurgical wound infections accounted for ∼60% of nmMRSA and MSSA SSTIs (115 of 198 infections and 242 of 390 infections, respectively), whereas surgical site infections made up 21 (51%) of 41 mMRSA SSTIs. When colonized individuals were excluded, sepsis, severe sepsis, and septic shock combined accounted for 52 (23%) of 225 nmMRSA infections, 108 (25%) of 436 MSSA infections, and 23 (39%) of 59 mMRSA infections. There was no statistically significant difference between nmMRSA and MSSA when comparing the severity of illness (P=.49), the distribution of overall clinical syndromes (P=.78), or the clinical syndromes in the SSTI group alone (P=.12)
OutcomesThe majority of patients were admitted to the hospital, and ∼50% of patients who had nmMRSA or MSSA isolated required surgical intervention (120 of 239 patients and 251 of 478 patients, respectively). The median length of stay was short for patients who had nmMRSA isolated (4 days) or MSSA isolated (3 days) but prolonged for patients who had mMRSA isolated (28 days) cases. The in-hospital mortality rate was higher for patients who had mMRSA isolated (table 1) than for the other groups
Antimicrobial susceptibilityDisk-approximation testing of erythromycin-resistant isolates revealed that inducible clindamycin resistance was present in 52 (22%) of 239 nmMRSA isolates and 77 (16%) of 478 MSSA isolates, respectively. All nmMRSA and MSSA isolates were susceptible to trimethoprim-sulfamethoxazole and rifampin, and most were susceptible to tetracycline (233 nmMRSA isolates [97%] and 475 MSSA isolates [99%]) and fusidic acid (206 [86%] nmMRSA isolates and 456 [95%] MSSA isolates). Patients who had nmMRSA isolated were less likely than those who had MSSA or mMRSA isolated to receive an antibiotic that was active against the causative organism (i.e., active antibiotic). The active antibiotics most commonly prescribed as inpatient therapy for nmMRSA infection were vancomycin (prescribed for 43 [54%] of 79 patients) and trimethoprim-sulfamethoxazole (prescribed for 49 [62%] of 79 patients)
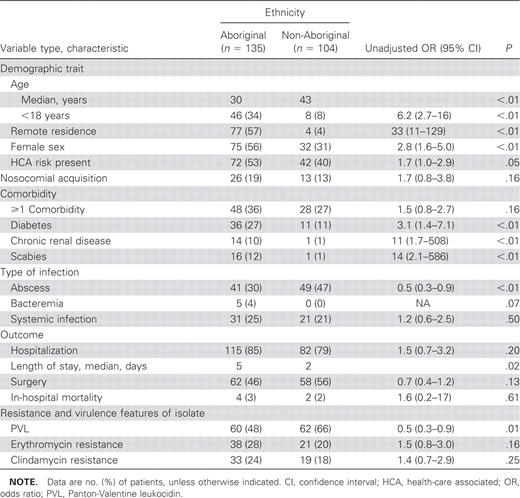
Comparison of Aboriginal and non-Aboriginal patients who had nmMRSA isolatedAboriginal patients who had nmMRSA isolated were significantly younger, more likely to be from remote regions, and less likely to be male than non-Aboriginal patients who had this organism isolated (table 2). Despite a younger age, diabetes mellitus and chronic renal disease were more common in Aboriginal patients and the length of stay was longer. Scabies was diagnosed 14 times more frequently in Aboriginal patients. Non–multidrug-resistant MRSA isolates from Aboriginal patients were less likely to be pvl positive
Characteristics of 239 patients from whom non–multidrug-resistant, methicillin-resistant Staphylococcus aureus isolated, stratified by ethnicity
Multivariate analysisBecause the characteristics of patients who had mMRSA isolated were clearly different from those of patients who had nmMRSA or MSSA isolated, we concentrated on comparing predictors for the isolation of nmMRSA to predictors for the isolation of MSSA. After multivariate conditional logistic regression, older age (adjusted OR [aOR] for each additional year, 1.01 [95% CI, 1.00–1.02]), female sex (aOR, 1.5 [95% CI, 1.1–2.0]), and remote residence (aOR, 1.8 [95% CI, 1.2–2.5]) predicted isolation of nmMRSA. Among patients who had nmMRSA isolated, Aboriginal patients were younger (aOR for each additional year, 0.94 [95% CI, 0.92–0.96]), more likely to be female (aOR, 3.8 [95% CI, 1.7–8.5]), more likely to reside remotely (aOR, 29 [95% CI 8.9–94]), more likely to have chronic renal impairment (aOR, 42 [95% CI, 4.5–403]), and less likely to be infected with a pvl-positive strain (aOR, 0.2 [95% CI, 0.1–0.6]) than non-Aboriginal patients
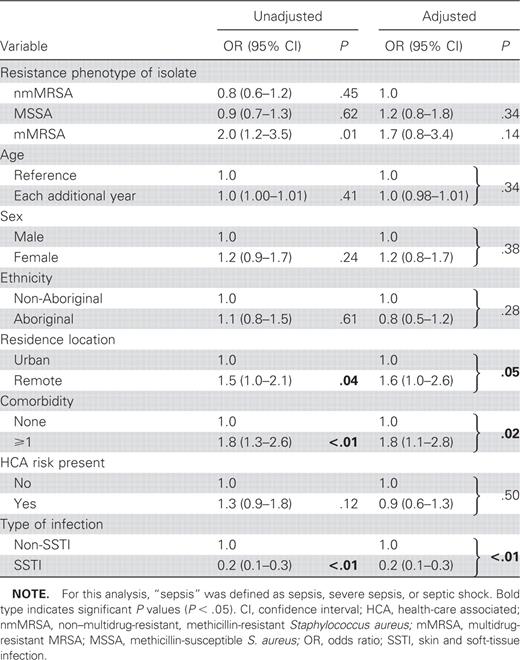
After multivariate logistic regression comparing sepsis (sepsis, severe sepsis, or septic shock) with localized infection only, the presence of comorbidities (aOR, 1.8 [95% CI, 1.1–2.8]) and remote residence (aOR, 1.6 [95% CI, 1.0–2.6]) predicted sepsis. SSTIs were associated with localized infection (aOR, 0.2 [95% CI, 0.1–0.3]). The isolate’s methicillin-resistance phenotype was not associated with severity of disease (table 3). For nmMRSA, the presence of pvl was the only factor significantly associated with sepsis (aOR, 2.2 [95% CI, 1.1–4.6])
Multivariate analysis of variables associated with localized infection versus sepsis
Discussion
The present study has revealed a heavy burden of staphylococcal disease in Aboriginal populations and defined the epidemiological and clinical relationship between nmMRSA and MSSA in northern Australia. The findings that patients who had nmMRSA isolated were significantly less likely to be male than those who had MSSA isolated and that remote residence was a strong predictor of nmMRSA isolation provide important insights into the emergence of nmMRSA. Evidence that clinical disease presentation and outcomes were independent of resistance phenotype suggests that, in our setting, virulence is not convergent with resistance phenotype
The incidences of both MSSA infection and nmMRSA infection are much higher in Aboriginal populations. These incidence rates strongly correlate with measures of remoteness and socioeconomic disadvantage across regions of the Top End. The annual incidence of S. aureus bacteremia—65 cases per 100,000 population—is almost double the rate estimated for the whole of Australia, which is 35 cases per 100,000 population [23] and more than double the rates recently reported from Canada [24] and Sweden [25]. When patients were stratified by ethnicity, the non-Aboriginal population of the Top End had an annual incidence of bacteremia similar to that of the rest of Australia but the incidence among the Aboriginal population is 6 times higher, at 172 cases per 100,000 population
Clinical and epidemiological risk factors do not appear to reliably distinguish between infection due to CA-MRSA strains and infection due to MSSA strains [26, 27], and outcomes following hospital discharge are comparable [28]. In Taiwan, there was no difference in the mortality rate for bacteremia due to CA-MRSA strains and that of bacteremia resulting from MSSA [29]. Our case-control study also demonstrated that nmMRSA and MSSA cause similar infections in a comparable group of patients. Similarities existed with respect to disease spectrum and outcomes, as well as most demographic and epidemiological features. This was in marked contrast to the results for mMRSA, which was isolated from a clearly distinct group of patients and resulted in different infections and outcomes
Multivariate analysis revealed 2 striking differences between patients who had nmMRSA isolated and those who had MSSA isolated. Our finding that those who had nmMRSA isolated were more likely to be female, or rather, less likely to be male, is surprising and intriguing. Previous studies with a comparable design [14, 26, 27, 30, 31] have consistently shown a predominance of males among patients with staphylococcal infections, which, to our knowledge, remains unexplained. After stratifying patients who had nmMRSA isolated according to ethnicity, we found that females comprised more than half of the Aboriginal patients but less than a third of non-Aboriginal patients; this latter figure mirrored that of patients who had MSSA isolated across the study population, with 166 (35%) of 478 being female. Of note, Aboriginal patients were significantly younger, and children (age, <18 years) accounted for 46 (34%) of 135 Aboriginal patients, compared with 8 (8%) of 104 non-Aboriginal patients. Referral bias is unlikely to explain the observed differences in the sex of people requiring treatment in a major tertiary hospital. In our community setting, men with mild infections are less likely than women and children to present to community health clinics, but this is unlikely to be the case for severe infections. It has been postulated that infection with nmMRSA may be more dependent on skin-to-skin or fomite-to-skin transmission, rather than resulting from endogenous colonization [32]. It has also been demonstrated that infants and mothers often share strains of S. aureus [33]. In Aboriginal communities, domestic crowding—as many as 7.5 people per bedroom—facilitates the transmission of infectious agents within households [34], and women provide most of the child care. Thus women, through their closer household contact with children, may be more likely to develop nmMRSA infection than men
Although both MSSA and nmMRSA are more common among Aboriginal people than non-Aboriginal people in the Top End, we have found that remote residence, rather than ethnicity itself, is most strongly associated with infection due to nmMRSA rather than MSSA. This contrasts with the results of studies from the United States, in which ethnicity was associated with nmMRSA infection [30, 31, 35, 36]. The remote areas of the Top End include Aboriginal communities characterized by socioeconomic disadvantage in relation to health, housing, education, and employment [18]. Because women and children were overrepresented among Aboriginal patients who had nmMRSA isolated, our findings support the postulate that factors in remote Aboriginal communities, such as domestic crowding and poor hygiene as well as high rates of scabies, skin sores, and antibiotic use, may contribute to the transmission and emergence of nmMRSA [13]. In addition, children in remote communities repeatedly present to community clinics with respiratory tract and skin infections [37] and are typically treated with penicillin and cephalosporin antibiotics
The methicillin-resistance phenotype did not predict whether an infection was localized or led to sepsis, and it was not predictive of the clinical spectrum of disease. Unlike the situation in the United States, where virulence and resistance have converged in the predominant USA300 clone [38], there continues to be a diversity of nmMRSA strains circulating in Australia [12]. Our current study and another from the Australian state of Queensland have demonstrated that virulence does not correlate with resistance phenotype in these settings [14]. We have previously established that nmMRSA and MSSA strains from the same clonal complexes circulate in remote Top End communities [10], suggesting that staphylococcal chromosome cassette (SCC) mecIV has independently been acquired by MSSA on multiple occasions in our environment. It has been demonstrated that the deletion of SCCmecIV in an isogenic mutant of a clinical USA300 isolate had no effect on competitive fitness or virulence [38]. It is therefore unsurprising that we found the clinical spectrum and outcomes of disease due to nmMRSA and MSSA to be similar
Interestingly, the presence of pvl genes in nmMRSA was associated with double the odds of sepsis. There continues to be debate over the importance of Panton-Valentine leukocidin (PVL), and mouse infection models have not consistently shown a major role in pathogenesis [39]. A recent clinical study found no significant difference between PVL-positive and PVL-negative USA400 isolates with respect to their propensity to cause infection or colonization [40]. However, there were only 10 PVL-positive isolates in that study, and there was a trend linking the presence of PVL with increasing severity of disease. Our findings support previous studies that demonstrated that patients with pneumonia or bone and joint infections caused by PVL-positive S. aureus were systemically more unwell that those with infections caused by PVL-negative S. aureus [41, 42]
nmMRSA has been present in northern Australia and the RDH since at least the early 1990s [4, 43]. Over the past 10–15 years, the percentage of nmMRSA and mMRSA isolates among the overall population of S. aureus isolates has increased from 4% [4] to 291 (17%) of 1693 for nmMRSA and from 3% [4] to 137 (8%) of 1693 for mMRSA. However, nmMRSA has not outstripped mMRSA as a nosocomial pathogen at the RDH. In 1991–1995, nmMRSA constituted 40% [43] of nosocomial MRSA isolates, compared with 39 (41%) of 95 in this 2006–2007 case-control study. The finding that nmMRSA and MSSA infection had similar clinical profiles and outcomes calls into question current infection control strategies that target MRSA on the basis of methicillin-resistance phenotype alone. It is possible that the spread of a potentially more virulent, PVL-positive MSSA strain may have more significant consequences than the spread of a PVL-negative nmMRSA strain, and therefore, it should be targeted for more aggressive infection control interventions. Molecular diagnostic methods may facilitate a different approach. One Australian state, Western Australia, has recently attempted to implement a “search and destroy” approach to PVL-positive MRSA strains in the community [44]; this extends existing policies of public health agencies being notified of all MRSA isolations and all MRSA isolates being sent to a reference typing laboratory
Our study has some limitations. We did not interview patients directly and were unable to assess the impact of factors such as domestic crowding, prior antibiotic use, and the mode of acquisition for SSTIs. Our incidence figures and case-control study pertain only to patients presenting to hospitals and do not reflect the overall community burden of staphylococcal infections. Nevertheless, these hospitals service an area of 510,000 km2, and our study provides important insights into staphylococcal infections in this large tropical region. Additionally, our calculations are likely to result in an underestimation of the already high incidence of S. aureus infection. Among children in some remote communities, the prevalence of pyoderma is 20%, from which S. aureus can be cultured in 57% of cases [10]. Hence, in these communities the prevalence of staphylococcal infection among children is >11,000 cases per 100,000 population, and the annual incidence of all staphylococcal infections is even higher
Staphylococcal disease, including that due to nmMRSA, imposes a disproportionate burden on remote Aboriginal communities in northern Australia. Our study demonstrates that female sex and residence in a remote community is associated with nmMRSA infection, and remote residence is likely a marker of social disadvantage that encompasses overcrowding, poor housing conditions, and lack of access to the physical infrastructure needed to maintain skin hygiene. There are well-founded fears that nmMRSA will become a nosocomial pathogen, but primary prevention also requires consideration of public health issues, such as housing and community hygiene. In our situation, nmMRSA is still principally a community pathogen—arising and spreading from within the community setting
Acknowledgments
We thank Paul Southwell, Gary Lum, Desmond Chih, and the Northern Territory Government Pathology Service Microbiology Laboratory for identifying and collecting isolates. We thank Elizabeth Canale, Marianne Martinello, and Jonas Zehnder for assistance in collecting patient data. We also thank Wendy Munckhof and Graeme Nimmo for assistance in the study design and Miles Beaman for data from the local private pathology service
References
(See the editorial commentary by Turnidge, on pages 1416–8.)
Potential conflicts of interest: none reported
Presented in part: 47th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, 17–20 September 2007, Chicago, Illinois (abstract L1141)
Financial support: The Healthy Skin Project received funding from the Australian National Health and Medical Research Council (S.T. is a PhD scholar; NHMRC grant 436033) and the Australian Cooperative Research Centre for Aboriginal Health