-
PDF
- Split View
-
Views
-
Cite
Cite
Mohammad Pouryasin, Heidar Sharafi, Bita Behnava, Seyed Moayed Alavian, Maryam Keshvari, Ali Pouryasin, A Simple PCR-RFLP Method for Genotyping of IFNL4 rs368234815 Polymorphism in Patients With Chronic Hepatitis C, Laboratory Medicine, Volume 48, Issue 1, February 2017, Pages 51–56, https://doi.org/10.1093/labmed/lmw060
Close - Share Icon Share
Background: The IFNL4 rs368234815 polymorphism plays a prominent role in spontaneous and treatment-induced clearance of hepatitis C virus (HCV) infection. This study aimed to develop a polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method for assessment of rs368234815 polymorphism.
Methods: We genotyped the rs368234815 polymorphism in 87 patients with chronic HCV by PCR sequencing and PCR-RFLP methods, simultaneously.
Results: Genotyping of IFNL4 rs368234815 via PCR-RFLP was concordant with PCR sequencing in all 87 individuals (100%). The analytical sensitivity and specificity of the developed PCR-RFLP method for genotyping of rs368234815 polymorphism were each 100%. Among these patients with chronic HCV, the frequency of rs368234815 TT/TT, TT/ΔG, and ΔG/ΔG were 44.8%, 37.9%, and 17.3%, respectively.
Conclusions: The PCR-RFLP method that we developed is accurate, rapid, inexpensive, and easy to perform for genotyping of the IFNL4 rs368234815 polymorphism. This method can be used for clinical and research work.
Worldwide, approximately 150 million people are infected with hepatitis C virus (HCV), and almost 500,000 people die annually from chronic HCV infection–related illnesses such as cirrhosis and hepatocellular carcinoma.1 In recent years, HCV treatment was tied to combination therapy consisting of pegylated interferon (PEG-IFN) and ribavirin (RBV).2‐4 Recently, new antiviral agents known as direct-acting antivirals (DAAs) have been developed and introduced for treatment of HCV infection.5,6 Although DAAs are more effective than PEG-IFN/RBV combination therapy, given these new treatments of HCV are not affordable and available in many countries, PEG-IFN and RBV are colloquially known to remain the alternative HCV treatment regimen.
Recently, studies7‐11 have shown that response to PEG-IFN/RBV combination therapy has been affected by host and virus genetic factors. Among the host genetic factors, rs368234815 (ss469415590) polymorphism within exon-1 of the interferon lambda 4 (IFNL4) gene (HGNC:44480) plays a prominent role in spontaneous and treatment-induced clearance of HCV infection.9 The IFNL4 gene, with a 2543 base-pair (bp) length and 5 exons, is located on chromosome 19q13.2. The rs368234815 polymorphism is classified as a deletion/insertion genetic variation (TT/ΔG). The IFNL4 rs368234815-ΔG allele expresses the IFN-λ4 protein; however, the rs368234815-TT allele cannot express the functional IFN-λ4 protein as a result of frameshift polymorphism that introduces a premature stop codon that blocks the correct protein translation.9 The results of the studies have shown that patients with the rs368234815-TT/TT genotype have a greater chance of favorable response to PEG-IFN/RBV treatment, compared with patients with rs368234815-TT/ΔG and -ΔG/ΔG genotypes. Similar results also have been observed in spontaneous clearance of HCV infection.12 The results of previous studies9,13‐25 determined the IFNL4 rs368234815 genotype using methods that require costly instruments and materials. Development of a simple, inexpensive, and rapid method for genotyping of IFNL4 rs368234815 polymorphism might be helpful. The aim of this study was to develop a simple, inexpensive, and rapid polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method for genotyping of IFNL4 rs368234815 polymorphism.
Material and Methods
Study Population
In this study, our cohort consisted of 87 patients with chronic HCV infection (70 males and 17 females), with a mean (SD) age of 30.6 (9.8) years, who were treated at Tehran Hepatitis Center and whose specimens were referred to the Armin Pathobiology Laboratory (both institutions are located in Tehran, Iran). The criteria for chronicity of HCV infection were having positive results for HCV antibodies and HCV RNA for a period of longer than 6 months. All study participants provided written informed consent; the study design was approved by the ethics committee of Baqiyatallah Research Center for Gastroenterology and Liver Diseases. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
IFNL4 Genotyping by PCR Sequencing
Genomic DNA was extracted from 200 µL of buffy coat fluid using the QIAamp DNA Blood Mini Kit (QIAGEN) according to manufacturer instructions. The direct DNA sequencing method for assessment of IFNL4 rs368234815 polymorphism was described previously.26 Briefly, we performed PCR reactions using Accupower PCR PreMix (Bioneer Corporation). We amplified 100 ng to 300 ng of genomic DNA using 10 pmol of rs12-F and rs12-R primer pairs (Table 1). The PCR temperature profile was as follows: 94 °C for 5 minutes, 35 cycles of 94 °C for 20 seconds, 66 °C for 20 seconds, and 72 °C for 20 seconds, followed by 72 °C for 5 minutes.
Primers for PCR Sequencing and PCR-RFLP Genotyping of the rs368234815 Polymorphism
| Method . | Primer Name . | Sequence . | GRCh38.p2 Primary Assembly No. . | TM(°C)a . | PCR Product Size (bp) . |
|---|---|---|---|---|---|
| PCR sequencing | rs12-F | 5′-GCGGAAGGAGCAGTTGCGCT-3′ | 39247951-39247970 | 66.08 | 745/746 |
| rs12-R | 5′-GTGCCTTCACGCTCCGAGCA-3′ | 39248677-39248696 | 65.46 | ||
| PCR-RFLP | IFNL4-F | 5′-GACGCAGGACCCCTTGGGACAGGA-3′ | 39248340-39248363 | 69.89 | 226/227 |
| IFNL4-R | 5′-TCTGGGCCGCAGTGGCCGCGAGG-3′b | 39248544-39248566 | 75.38 |
| Method . | Primer Name . | Sequence . | GRCh38.p2 Primary Assembly No. . | TM(°C)a . | PCR Product Size (bp) . |
|---|---|---|---|---|---|
| PCR sequencing | rs12-F | 5′-GCGGAAGGAGCAGTTGCGCT-3′ | 39247951-39247970 | 66.08 | 745/746 |
| rs12-R | 5′-GTGCCTTCACGCTCCGAGCA-3′ | 39248677-39248696 | 65.46 | ||
| PCR-RFLP | IFNL4-F | 5′-GACGCAGGACCCCTTGGGACAGGA-3′ | 39248340-39248363 | 69.89 | 226/227 |
| IFNL4-R | 5′-TCTGGGCCGCAGTGGCCGCGAGG-3′b | 39248544-39248566 | 75.38 |
TM, Melting Temperature; PCR, polymerase chain reaction; PCR-RFLP, polymerase chain reaction–restriction fragment length polymorphism; bp, base pair.
aTM calculation of primers was performed using the SantaLucia formula.37
bThe underlined nucleotide represents mismatch, to abolish the nonspecific restriction site near the rs368234815 polymorphism.
Primers for PCR Sequencing and PCR-RFLP Genotyping of the rs368234815 Polymorphism
| Method . | Primer Name . | Sequence . | GRCh38.p2 Primary Assembly No. . | TM(°C)a . | PCR Product Size (bp) . |
|---|---|---|---|---|---|
| PCR sequencing | rs12-F | 5′-GCGGAAGGAGCAGTTGCGCT-3′ | 39247951-39247970 | 66.08 | 745/746 |
| rs12-R | 5′-GTGCCTTCACGCTCCGAGCA-3′ | 39248677-39248696 | 65.46 | ||
| PCR-RFLP | IFNL4-F | 5′-GACGCAGGACCCCTTGGGACAGGA-3′ | 39248340-39248363 | 69.89 | 226/227 |
| IFNL4-R | 5′-TCTGGGCCGCAGTGGCCGCGAGG-3′b | 39248544-39248566 | 75.38 |
| Method . | Primer Name . | Sequence . | GRCh38.p2 Primary Assembly No. . | TM(°C)a . | PCR Product Size (bp) . |
|---|---|---|---|---|---|
| PCR sequencing | rs12-F | 5′-GCGGAAGGAGCAGTTGCGCT-3′ | 39247951-39247970 | 66.08 | 745/746 |
| rs12-R | 5′-GTGCCTTCACGCTCCGAGCA-3′ | 39248677-39248696 | 65.46 | ||
| PCR-RFLP | IFNL4-F | 5′-GACGCAGGACCCCTTGGGACAGGA-3′ | 39248340-39248363 | 69.89 | 226/227 |
| IFNL4-R | 5′-TCTGGGCCGCAGTGGCCGCGAGG-3′b | 39248544-39248566 | 75.38 |
TM, Melting Temperature; PCR, polymerase chain reaction; PCR-RFLP, polymerase chain reaction–restriction fragment length polymorphism; bp, base pair.
aTM calculation of primers was performed using the SantaLucia formula.37
bThe underlined nucleotide represents mismatch, to abolish the nonspecific restriction site near the rs368234815 polymorphism.
PCR products were sequenced using BigDye Terminator V3.1 Cycle Sequencing Kits (Applied Biosystems Inc.) in a 3130XL ABI Genetic Analyzer (Applied Biosystems Inc.) according to manufacturer instruction. Sequencing results were analyzed using Chromas Lite Version 2.1.1 (Technelysium).
IFNL4 Genotyping via PCR-RFLP
We used the IFNL4-F and IFNL4-R primers as a forward and reverse primer pair (Table 1). The PCR reactions were performed using Accupower PCR PreMix. The DNA amount and primer concentrations for the PCR mixture were the same as in the PCR sequencing method. The PCR temperature profile was as follows: 94 °C for 5 minutes, 40 cycles of 94 °C for 20 seconds, 60 °C for 30 seconds, and 72 °C for 30 seconds, followed by 72 °C for 5 minutes. For the RFLP analysis, the PCR product of rs368234815 was digested with 10 units of MspA1I (New England BioLabs) restriction endonuclease (RE) for more than 1 hour. The digested PCR products were separated on 3% agarose gel alongside the GeneRuler 100-bp DNA Ladder (Thermo Fisher Scientific Inc.). The agarose gel was stained with the addition of 0.5 µL of DNA safe Stain (CinnaGen Co.) into each 100 mL of agarose gel. In each run of PCR-RFLP genotyping of rs368234815, as a control for enzymatic activity, we included a single rs368234815-ΔG/ΔG DNA specimen.
Validation Protocol
For the validation of the PCR-RFLP method for genotyping of IFNL4 rs368234815, the PCR-RFLP genotyping results should be 100% concordant with the DNA sequencing results.
Results
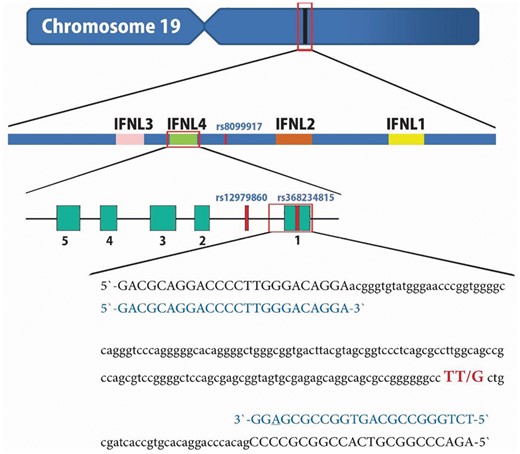
IFNL4 Genomic Region and the PCR-RFLP Primer Design
Chromosome 19, IFNL4 gene, rs368234815 polymorphism and the primers for polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP).
Finally, based on the aforementioned findings, we designed the PCR primers based on 2 goals. First, we aimed to give consideration to the homologous region; second, we aimed to abolish nonspecific RS, which is located downstream of the IFNL4 rs368234815 polymorphism. We attempted to design the primers restricted to regions with the minimum homology. Further, in the reverse primer, we chose a conventional mismatched nucleotide near the 3′ end of the primer, to abolish the nonspecific RS (Table 1).
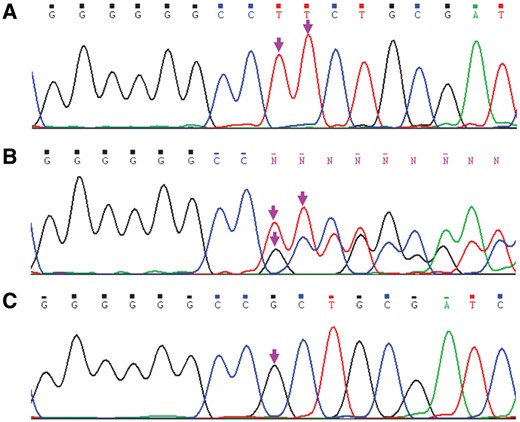
PCR Sequencing Set-Up
PCR sequencing chromatograms of rs368234815 polymorphism. A, Sequencing chromatogram of IFNL4 rs368234815-TT/TT genotype. B, Sequencing chromatogram of IFNL4 rs368234815-TT/ΔG genotype. C, Sequencing chromatogram of IFNL4 rs368234815-ΔG/ΔG. Arrows show the polymorphic sites.
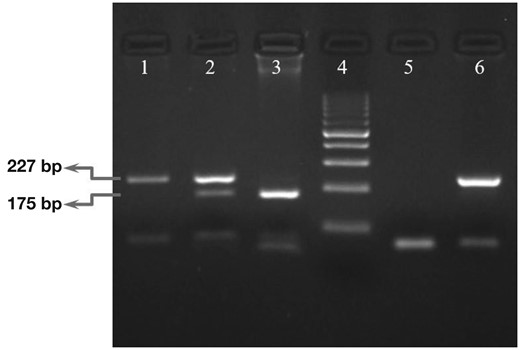
PCR-RFLP Set-Up
Polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) electrophoresis results of IFNL4 rs368234815 polymorphism. Lanes 1, 2, and 3 were genotyped as TT/TT, TT/ΔG and ΔG/ΔG, respectively, for IFNL4 rs368234815. Lane 4 was a 100-bp ladder. Lanes 5 and 6 were no template control and nondigested PCR product, respectively. For electrophoresis of digested polymerase chain reaction (PCR) products, 3% agarose gel was used.
Validation Results
The results of genotyping of the rs368234815 polymorphism by PCR-RFLP were concordant with those of PCR sequencing in all 87 individuals (100%) and showed the 100% analytical sensitivity and specificity of the PCR-RFLP method. In both methods, the frequencies of TT/TT, TT/ΔG, and ΔG/ΔG were 44.8%, 37.9%, and 17.3%, respectively.
Discussion
The final goal of HCV treatment is the eradication of the virus, which also can be determined by sustained virologic response (SVR), or a negative result for HCV RNA 6 months or longer after treatment termination.2,4 Several genetic markers have been proposed as the predictors of SVR, including HCV and host genetic factors. In 2009, 3 genome-wide association studies7,29,30 identified a genomic region located upstream of IFNL3 (formerly IL28B) that contains several single-nucleotide polymorphisms (SNPs) associated with the rate of SVR to treatment with PEG-IFN/RBV. These markers are in strong linkage disequilibrium (LD) with each other. The results of further studies31‐33 have shown that these markers are also associated with spontaneous clearance of HCV infection. Among these markers, IFNL3 rs12979860 polymorphism has attracted the most attention for testing as a pretreatment marker, for prediction of response to PEG-IFN/RBV combination therapy.34 The studies attempted to identify the functional mechanisms linked to IFNL3 SNPs, but those efforts failed.35
In 2013, sequencing of RNA specimens from primary hepatocytes that underwent treatment with polyinosinic:polycytidylic acid (a synthetic double-stranded RNA that mimics HCV infection) uncovered a novel gene called IFNL4, which is located upstream of the IFNL3 gene.9,IFNL4 contains rs368234815 within exon-1 and rs12979860 within intron-1. These 2 polymorphisms are linked together in ethnic Asian and ethnic European populations; less LD was observed in ethnic Africans. Evaluations of African American patients with HCV infection indicated that rs368234815 may be the strongest host genetic factor for prediction of HCV treatment response.12,36 Designing a simple, inexpensive, and rapid method for genotyping of the IFNL4 rs368234815 polymorphism might be helpful for predicting response to treatment. Further, new studies, which will require the determination of IFNL4 rs368234815 genotypes, can be easily performed using the PCR-RFLP method, even in local centers.
Until now, several studies9,15,16,19,21‐24 have genotyped IFNL4 rs368234815 with probe-based assays, including the TaqMan and Invader assays.13,20 Wu et al25 genotyped rs368234815 using the mass spectrometry method. Keshvari et al17 used the PCR sequencing method to genotype the IFNL4 rs368234815 polymorphism. Galmozzi et al14 and Lamoury et al18 detected IFNL4 rs368234815 genotype using the high-resolution melting (HRM) assay.
Overall, these methods have certain limitations, including dependence on instruments with sophisticated technology and possible lack of affordability by some laboratory centers. Probe-based and HRM assays can be used as high-throughput methods, and the testing procedures can be acceptably speedy for a large number of specimens. Although the present PCR-RFLP method is not acceptable as a high-throughput method, it can serve as a high-speed method for genotyping of a small number of specimens. The present method and almost all of the aforementioned methods require a control specimen for assessment of the validity of the testing procedures. In the present study, we used an IFNL4 rs368234815-ΔG/ΔG specimen as the digestion control. Despite the advantages of probe-based assays, the probes involved are sensitive; synthesis of those probes requires instruments with sophisticated technology. The present study is primer-dependent; synthesis of the primers can be performed even by local providers. In the PCR sequencing method, the PCR step is primer-dependent; however, further steps, including the sequencing step, require instruments that feature sophisticated technology, which may be costly compared with the PCR-RFLP method.
In conclusion, the PCR-RFLP method that we have developed for genotyping of rs368234815 is accurate, inexpensive, rapid, and easy to perform. This method is comparable to the PCR sequencing method, which is the criterion-standard method for detecting genetic markers, and so it can be used for clinical and research work. LM
References
World Health Organization (WHO). Hepatitis C. Geneva, Switzerland: World Health Organization;
Abbreviations
- HCV
hepatitis C virus
- PEG-IFN
pegylated interferon
- RBV
ribavirin
- DAAs
direct-acting antivirals
- IFNL4
interferon lambda 4
- bp
base pair
- PCR-RFLP
polymerase chain reaction–restriction fragment length polymorphism
- RE
restriction endonuclease
- IFNL2
interferon lambda 2
- IFNL3
interferon lambda 3
- RS
restriction site
- SVR
sustained virologic response
- SNPs
single nucleotide polymorphisms
- LD
linkage disequilibrium
- HRM
high resolution melting
- TM
Melting Temperature