-
PDF
- Split View
-
Views
-
Cite
Cite
Shuai Weng, Qing Dong, Rama Balakrishnan, Karen Christie, Maria Costanzo, Kara Dolinski, Selina S. Dwight, Stacia Engel, Dianna G. Fisk, Eurie Hong, Laurie Issel-Tarver, Anand Sethuraman, Chandra Theesfeld, Rey Andrada, Gail Binkley, Christopher Lane, Mark Schroeder, David Botstein, J. Michael Cherry, Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins , Nucleic Acids Research, Volume 31, Issue 1, 1 January 2003, Pages 216–218, https://doi.org/10.1093/nar/gkg054
Close - Share Icon Share
Abstract
The Saccharomyces Genome Database (SGD: http://genome-www.stanford.edu/Saccharomyces/ ) has recently developed new resources to provide more complete information about proteins from the budding yeast Saccharomyces cerevisiae . The PDB Homologs page provides structural information from the Protein Data Bank (PDB) about yeast proteins and/or their homologs. SGD has also created a resource that utilizes the eMOTIF database for motif information about a given protein. A third new resource is the Protein Information page, which contains protein physical and chemical properties, such as molecular weight and hydropathicity scores, predicted from the translated ORF sequence.
Received September 13, 2002; Accepted September 24, 2002
INTRODUCTION
The Saccharomyces Genome Database (SGD) collects, organizes, and presents information about the molecular biology and genetics of the budding yeast Saccharomyces cerevisiae . SGD contains diverse types of biological data and provides tools for their search and analysis. Information in SGD is generally organized around a gene; each gene in the genome has a Locus page ( 2 ), which contains basic information about that gene such as Gene Ontology (GO) annotations ( 1 , 3 ) and phenotype data as well as links to additional tools and resources. The protein resources described below are newly provided links located in the Protein Info and Structure Resources section of the Locus page.
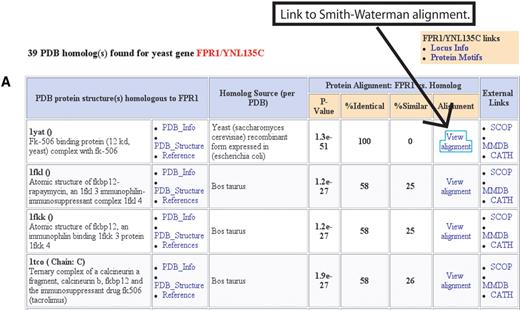
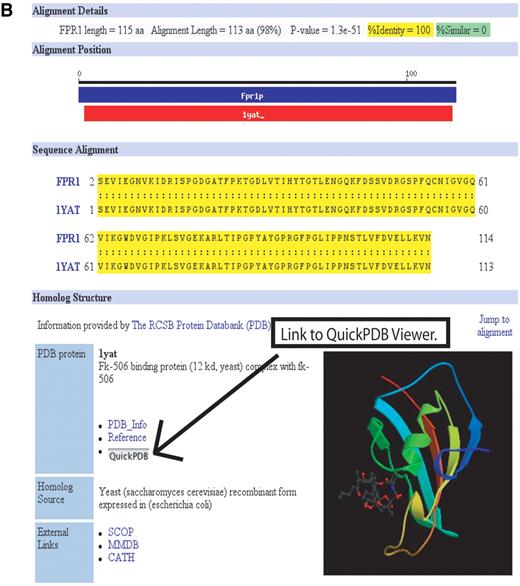
The Protein Data Bank (PDB) Homologs page provides a list of structures available in PDB ( 7 ) relevant to a given S. cerevisiae protein (Fig. 1 A). The list is generated by comparing the sequences of the systematically defined S. cerevisiae proteins against the protein sequences within PDB using the Smith–Waterman ( 6 ) sequence comparison algorithm. All PDB sequences with a p -value of 0.01 or less are presented, regardless of species. Thus, if the structure of an S. cerevisiae protein is unknown, the structure of the homo-logous protein in another species may be available. For each PDB homolog, Smith–Waterman scores are provided, with links to PDB, other protein databases, and an alignment page. The alignment page (Fig. 1 B) displays the Smith–Waterman alignment of the S. cerevisiae protein against the PDB protein, a colour ribbon image of the structure, and a link to the interactive QuickPDB structure viewer provided by PDB.
SGD has recently released another protein information tool. The new Motif tool displays the motifs in a particular protein using results from the eMOTIF resource ( 4 ). The Motif results page includes an image illustrating all the motifs in the protein of interest and also lists all the other S. cerevisiae proteins that share these motifs. Links to other motif databases are also provided.
The third new protein resource in SGD is the Protein Information page. This page contains calculated data based on protein sequence, such as molecular weight, amino acid content, protein length, pI, and codon adaptation index ( 5 ). Links are provided to graphical displays of hydropathy plots, helical wheels and other tools to predict secondary structure. SGD is developing enhancements to this page and will soon be incorporating predicted transmembrane domains, signal sequences and other types of data that can be generated based on protein sequence. All of these data for the entire predicted yeast proteome are available for download on the SGD anonymous FTP site at: ftp://genome-ftp.stanford.edu/yeast/data_download/protein_info/ .
From its beginning in 1994, SGD has provided biological information about yeast genes as well as search and analysis tools. We are now expanding our scope to provide more protein information by developing tools like the PDB Homolog, Motif, and Protein Information pages. SGD is also creating resources for global analyses, in addition to the gene-by-gene tools and resources currently provided. As more eukaryotic genomes are completely sequenced, comparative genomics will become an increasingly powerful method for solving biological problems; thus, SGD is exploring new ways of presenting the results of these and other types of genome-wide studies. Check the SGD home page at http://genome-www.stanford.edu/Saccharomyces/ for announcements as these new tools become available. Supplemental material for this paper can be found online at: http://genome-www.stanford.edu/Saccharomyces/help/NAR2003Supplement.html .
Figure 1. (Following page) Protein structure information at SGD: the new PDB Homologs tool. Due to space considerations, both ( A ) and ( B ) are just a portion of the web display. (A) The PDB Homologs results page lists the results of the Smith–Waterman sequence comparison of a S. cerevisiae protein against the proteins in PDB. Links are provided to the PDB and other external protein structure databases, as well as the alignment page shown in B. In this example, Fpr1p results are shown. A structure of the S. cerevisiae was identified (first row in the table), as well as additional structures of the Bos taurus homolog. Additional homologs were found but are not shown. This page can be reached from the ‘Protein Info and Structure’ pull-down menu on Locus and Protein Information pages. (B) The PDB Alignment page displays the alignment of the S. cerevisiae protein and the PDB protein, a color ribbon image of the structure (provided by PDB), and links to other databases and tools such as the interactive structural viewer QuickPDB provided by the PDB.
References
Ashburner,M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T., Harris,M.A., Hill,D.P., Issel-Tarver,L., Kasarskis,A., Lewis,S., Matese,J.C., Richardson,J.E., Ringwald,M., Rubin,G.M. and Sherlock,G. (
Ball,C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Issel-Tarver,L., Kasarskis,A., Scafe,C.R., Sherlock,G., Binkley,G., Jin,H., Kaloper,M., Orr,S.D., Schroeder,M., Weng,S., Zhu,Y., Botstein,D. and Cherry,J.M. (
Dwight,S.S., Harris,M.A., Dolinski,K., Ball,C.A., Binkley,G., Christie,K.R., Fisk,D.G., Issel-Tarver,L., Schroeder,M., Sherlock,G., Sethuraman,A., Weng,S., Botstein,D. and Cherry,J.M. (
Sharp,P.M. and Li,W.H. (
Smith,T.F. and Waterman,M.S. (




Comments