-
PDF
- Split View
-
Views
-
Cite
Cite
Roberta Sarno, Yoan Vicq, Norio Uematsu, Marine Luka, Clement Lapierre, Dana Carroll, Giacomo Bastianelli, Alexandre Serero, Alain Nicolas, Programming sites of meiotic crossovers using Spo11 fusion proteins, Nucleic Acids Research, Volume 45, Issue 19, 2 November 2017, Page e164, https://doi.org/10.1093/nar/gkx739
Close - Share Icon Share
Abstract
Meiotic recombination shapes the genetic diversity transmitted upon sexual reproduction. However, its non-random distribution along the chromosomes constrains the landscape of potential genetic combinations. For a variety of purposes, it is desirable to expand the natural repertoire by changing the distribution of crossovers in a wide range of eukaryotes. Toward this end, we report the local stimulation of meiotic recombination at a number of chromosomal sites by tethering the natural Spo11 protein to various DNA-binding modules: full-length DNA binding proteins, zinc fingers (ZFs), transcription activator-like effector (TALE) modules, and the CRISPR-Cas9 system. In the yeast Saccharomyces cerevisiae, each strategy is able to stimulate crossover frequencies in naturally recombination-cold regions. The binding and cleavage efficiency of the targeting Spo11 fusions (TSF) are variable, being dependent on the chromosomal regions and potential competition with endogenous factors. TSF-mediated genome interrogation distinguishes naturally recombination-cold regions that are flexible and can be warmed-up (gene promoters and coding sequences), from those that remain refractory (gene terminators and centromeres). These results describe new generic experimental strategies to increase the genetic diversity of gametes, which should prove useful in plant breeding and other applications.
INTRODUCTION
In eukaryotes, meiotic recombination between the homologous chromosomes disrupts the physical linkage between the parental alleles, generating the haplotype diversity transmitted to the offspring by the gametes. Recombination also helps homologous chromosome to pair and their physical connection by the crossover molecules, ensures accurate chromosome segregation during the first of the two meiotic divisions. The absence or the mis-localization of the crossovers is a source of sterility (1).
Meiotic recombination is neither randomly nor homogeneously distributed along the chromosomes, as reflected by the large distortion observed between the genetic and physical distances along the chromosomes as well as by the large variation in average recombination frequencies between species. Recombination is initiated by the evolutionarily conserved Spo11 protein, that generates DNA double-strand breaks (2,3), which are subsequently repaired on the homologous chromosomes to yield the non-crossover (gene conversion) and crossover molecules (CO) (4). In the yeast Saccharomyces cerevisiae, ∼160–200 DSBs form per cell and the majority leads to recombination events (5,6). In contrast, in Arabidopsis thaliana there are ∼200–300 DSBs per cell but only ∼10 crossovers occur per meiosis (7). Similarly, there are ∼200–300 DSBs per cell in mouse and only ∼24 crossovers per meiosis (8). Clearly, in every species, DSBs are more likely to occur in some genomic regions than in others, being related to the Spo11 chromatin association and cleavage probabilities varying by several order of magnitude from site to site but also due to the combinatorial action of several factors acting at different scales (5), including crossover homeostasis (9) and feedback mechanisms controlling DSB formation (10–12). Extreme is the regional distribution of crossovers towards the distal regions in barley and wheat crop species (13). For example, in the wheat chromosome 3B (∼1 Gb long), crossovers are clustered in only 13% of the chromosome (14). Thus, the highly skewed distribution of meiotic recombination generates large non-recombining regions and limits the ability to associate or separate genes over a small number of generations. To overcome such limit, the development of methods to program the frequency and localization of meiotic recombination remains to be achieved.
Mechanistically, meiotic recombination initiation is a ubiquitous and tightly regulated multi-step process, closely integrated with the development of meiosis-specific higher order chromosome structures (4,11,15–17). An important common determinant of DSB formation is the accessibility of the chromatin to the recombination machinery. Chromatin accessibility related to low nucleosome occupancy contributes to the bias that favours DSB formation and/or recombination in promoter regions as observed in yeasts (5,18) and A. thaliana (7). Nevertheless, not all nucleosome depleted regions are sites of DSB formation. Intriguingly, in contrast to the rapidly evolving PRDM9-dependent hotspots in mammals (19), recent studies in distant species of budding yeast (18) and birds (20) revealed that most hotspots are evolutionarily conserved, suggesting that strong mechanistic and/or evolutionary constraints play a role in designating the preferred DSB sites. In addition, the resolution of the recombination intermediates into gene conversion or crossover products is tightly controlled, limiting the number but ensuring at least one crossover per homolog.
Together, the complexity of the recombination process and its mechanistic and regulatory constraints suggested that it may be difficult to modify meiotic recombination frequencies and sites without inducing sterility. However, and perhaps paradoxically, meiotic recombination seems to remain flexible (17,21–24). Earlier experiments showed that, in yeast, fusion of Spo11 to the Gal4 DNA-binding domain (Gal4BD) was sufficient to induce DSB formation and recombination at Gal4 binding sites (Gal4UAS) in naturally cold regions (21,25,26). This Spo11-tethering strategy remains limited, being constrained to targeting endogenous Gal4UAS sites. In addition, many of these sites are not bound by the Gal4BD-Spo11 protein in vivo or are bound but remain refractory to DSB formation (26). In A. thaliana, where the total number of DSBs per cell greatly exceeds the number of crossovers, mutations or increased dosage of recombination modifiers (FANCM, FIGL1, RECQ4, HEI10) have led to an increase in recombination (27–30). However, these methods elevate crossover numbers solely in naturally initiating regions and does not allow to modify DSB refractory regions. Thus, to induce crossovers throughout the genome, it remains necessary to develop methods allowing to direct DSBs to arbitrarily chosen sites.
Here, we report the development of a series of targeting Spo11 fusions (TSFs), carrying a variety of DNA binding modules that allowed us to vary the location and/or the number of recombination initiation sites. Our results define a versatile and customizable Spo11-based platform to interrogate naturally DSB-cold regions and to target meiotic recombination in eukaryotic cells, highlighting its generic potential for biotechnological applications.
MATERIALS AND METHODS
Plasmid constructions
The plasmids used in this work are listed in Supplementary Table S1. All plasmids are derivatives of pAP1 and pAP11 (21,25) which contain the GAL4BD-SPO11 fusion gene flanked by the ADH1 promoter and transcription terminator. To exchange the GAL4BD region with other Spo11-targeting modules, the desired in vitro-built PCR fragment flanked by a SpeI and an XmaI site was introduced between the ADH1 promoter and the XmaI site located within the 36 nt internal linker (PEFMAMEAPGIR) that separates the targeting module and the N-terminal region of the full-length S. cerevisiae Spo11 coding region. Sequences encoding the full-length proteins Gal4 (560 aa), Tec1 (486 aa), Rsc3 (885 aa) and Mtw1 (290 aa) were amplified by PCR from the wild type SK1 yeast strain ORD7254–25D. The pRSM046, pRSM026, pRSM070 and pMLM023 plasmids contain the GAL4-SPO11, TEC1-SPO11, RSC3-SPO11 and MTW1-SPO11 fusions, respectively. To build the QQR-SPO11 gene construct, the QQR coding fragment (encoding 101 aa) was PCR-amplified from the QQR-pET15b plasmid (31) and cloned into the pAP11 plasmid to yield pAP119 plasmid. Regarding the dCAS9-SPO11 gene construct we first designed the sequence of a yeast codon-optimized version of the Cas9 protein from Streptococcus pyogenes (1368 aa) and containing inactivating D10A and H840A mutations (dCas9) (32). A nuclear localization signal (NLS, GGMAAPKKKRKVDGG) was attached to the N-terminus of dCas9 to promote nuclear import. The NLS and N-terminus of the dCas9 protein were separated by an internal linker (GIHGVPAA) (33). The NLS-dCAS9 construct was synthesized by GenScript and cloned into a pUC57 vector to create pScdCas9. The pRSM026 SpeI-XmaI fragment was replaced by the SpeI–XmaI fragment from pScdCas9 plasmid to yield the pAS502 plasmid. To attach the 6xHis-3xFlag tag to the C-terminus of the Spo11 protein, the 6xHis-3xFlag coding sequence was PCR amplified from ANT2152 (SPO11–6xHis-3xFlag-KanMX) and Gibson assembled into the NdeI-linearized pAS502 plasmid by using the NEB Gibson Assembly kit. The resulting pAS504 plasmid contains the PADH1-NLS-dCAS9-SPO11–6xHis-3xFlag-TADH1 construct. The dCas9-Spo11 and dCas9-Spo11–6xHis-3xFlag constructs display very similar capacities to complement spo11Δ sporulation defects and to induce meiotic DSB formation. The TALE array (encoding 1097 aa) was synthesized by GeneArt (Life Technologies™) and provided in the pDONR221 vector. The NLS-TALE fragment was cloned into a SpeI/XmaI-digested pRSM046 plasmid to obtain pMLM060. The DNA sequence of the seven TSF constructs is reported in the Supplementary Material (Figure S1).
The guide RNA (gRNA) expression plasmids were constructed from the PRPR1_gRNA_handle_TRPR1 plasmid (34) which was a gift from Timothy Lu (Addgene plasmid #49014). This plasmid, called thereafter p_gRNA_handle, encodes the gRNA backbone (without the guide sequence) whose expression is driven by the constitutive RPR1 promoter (an RNA-polymerase-III-dependent promoter). For the synthesis and cloning of each guide sequence into p_gRNA_handle, two complementary oligonucleotides, each carrying the 20-nt guide sequence, were designed. Both oligonucleotides also bear at the 5′ end a 40-nt sequence identical to the regions flanking the HindIII site of p_gRNA_handle. We used this pair of oligonucleotides to PCR amplify the 100-bp fragment and then cloned it into the HindIII site of the p_gRNA_handle vector by one-step Gibson assembly. Oligonucleotides used to construct each guide sequence are listed in Supplementary Table S2. To construct a multiplex gRNAs expression vector a multiple cloning sites sequence providing eight unique cloning sites (ApaI, NcoI, NdeI, PstI, SmaI, SphI, BamHI and BglII) was introduced into the SacI site of the p_gRNA_handle vector by one-step Gibson assembly, to yield the pAS508 plasmid. Multiple gRNA expression cassettes, each containing the RPR1 promoter, a gRNA sequence and the RPR1 terminator, were PCR amplified from the corresponding single gRNA expression plasmid and Gibson assembled into the NcoI site of the pAS08 plasmid. All plasmid constructs were verified by restriction digestion and Sanger sequencing.
Yeast strains
The genotypes of the diploid S. cerevisiae strains used in this work are listed in Supplementary Table S1. They are derivatives of SK1 (35) and were obtained by one-step transformation of haploid parents or recovered from genetic crosses. All strains, except ORD7304 (gift of V. Borde) and ORD8175 (25) were constructed for this work. For introduction into the yeast genome, all TSF expression plasmids except pAP119 plasmid, were linearized by digestion with XbaI. pAP119 plasmid was digested by Bsu36I. Linearized TSF plasmids were integrated at the TRP1 locus by transformation of the strain AND1940–4D (MATalpha, spo11::hisG-URA3-hisG, his4, leu2, trp1, ura3) or ORD7254–25D (MATa, his4, leu2, trp1, ura3) and verified by Southern blot analysis. gRNA expression plasmids were introduced into yeast cells by electroporation and transformants were selected on plates depleted in leucine. The sae2Δ, spo11Δ and gal4Δ mutations were introduced by genetic crossing using appropriate and previously described haploid strains (21).
Site-directed mutagenesis of the QQR RS located in the RSC9-MSC1 intergenic region was carried out using the Quick Change Site-directed Mutagenesis Kit (Stratagene) by mutating the GGG GAA GAA RS sequence into GGG GAT TAA, thus creating an AseI site. This mutation (iRSC9-MSC1 RS-AseI) was integrated into the yeast genome by two-step transformation and verified by Southern blot analysis.
The NatMX and HphMX resistant cassettes were PCR-amplified from plasmids pAG25 (36) and pMJ696 (identical to pAG32 (36)) and flanked by the yeast targeting regions of homology. To measure recombination frequencies around the GAL2 locus, the NatMX and HphMX cassettes were integrated in the terminator of GAL2 (GAL2ter) between positions 292068 and 292069 and in the promoter of EMP46 (EMP46prom) between positions 287725 and 287726 on chromosome XII, respectively. To examine the frequency of meiotic recombination near the PUT4 locus, NatMX and HphMX cassettes were inserted into the intergenic i(REV1-PYK2) and i(PUT4-CIN1) regions of chromosome XV at position 984842–984843 and 989657–989658, respectively. To study the effects of TEC1-SPO11 expression on meiotic recombination, the NatMX and HphMX cassettes were integrated in the promoter of the COX5A and VAC7 genes on chromosome XIV at positions 525932–525933 and 531522–531523, respectively. To examine the frequency of meiotic recombination around the HHT2-HHF2 region in chromosome XIV, the NatMX and HphMX cassettes were inserted between positions 574156 and 574157, and 578751 and 578752, respectively. To determine recombination frequencies around the MSC1 locus, the KanMX and HphMX markers were inserted into the intergenic i(PGA3-ERG13) and i(COX14-ERO1) regions at position 20682–20692 and 14249–14300 on chromosome XIII, respectively. The expected chromosomal integration sites were verified by junction PCR and Southern blotting.
Sporulation conditions
For meiotic time-courses, diploid cells were grown at 30°C in SPS presporulation medium and transferred into the sporulation medium (1% potassium acetate supplemented with amino acids) as previously described (37). Sporulation was performed at 30°C, 250 rpm. Tetrads were dissected after 96 h of sporulation.
Chromatin immunoprecipitation
ChIP experiments were performed as previously described (26). Briefly, about 8 × 108 meiotic CRISPR-dCAS9-SPO11 spo11Δ/spo11Δ sae2Δ/sae2Δ cells were treated with 1% formaldehyde for 15 min at room temperature and then 125 mM Glycin for 5 min. Immunoprecipitation was performed using 2 μg of the ANTI-FLAG mouse antibodies (Clone M2; Sigma-Aldrich) in combination with 35 μl of magnetic Dynabeads protein G (panMouse IgG, Dynal). Quantitation of immunoprecipitated DNA was performed via real-time quantitative PCR analysis, using Power SYBR Green mix (Applied Biosystems) and a 7900HT Fast Real-Time PCR System (Applied Biosystems). Quantitative PCR was performed on 1/60 of the immunoprecipitate or 1/20 000 of the whole-cell extract, and analyzed as described (37). To determine the non-specific background the amount of the SMC1 DSB-cold domain (38) was measured in each immunoprecipitate. Fold enrichments are expressed as the relative fold enrichment of target site over SMC1 signal and are normalized to input DNA. Sequence of the qPCR primers are listed in the Supplementary Table S1.
DSB mapping and quantification
Physical DSB detection was performed in strains homozygous for the sae2 allele (or the rad50S allele for QQR strains) to allow the accumulation of unrepaired DSBs. Genomic DNA was extracted as previously described (25). For Southern blot detection of 1 to 15 kb DNA fragments, genomic DNA or agarose-embedded DNA from meiotic samples was digested with appropriate restriction enzymes and the resulting fragments were separated by electrophoresis in 0.8–1.5% agarose gels and transferred under vaccum (Appligene) onto Hybond XL membranes (Amersham). DNA probes were PCR-amplified from genomic DNA and were labelled by 32P-dCTP using the Ready Prime kit (GE Healthcare). Probe hybridization of the membrane was performed as described previously (39). Parental and DSB bands were visualized by using a Typhoon 9400 Phospohoimager (GE Healthcare) and quantified with the ImageJ (NIH) software. DSB frequencies were measured as % of radioactivity in each DSB fragment relative to the total amount of radioactivity in the parental plus DSBs bands. For the majority of examined regions two independent experiments were performed to determine DSB frequencies. In these cases DSB values are the average of two experiments.
Yeast strains, restriction enzymes and probes are as follows:
Figure 2: genomic DNA was prepared from SPO11/SPO11 (ORD7304), GAL4-SPO11/GAL4-SPO11 spo11Δ/spo11Δ (AND2096), TEC1-SPO11/TEC1-SPO11 spo11Δ/spo11Δ (AND1926), RSC3-SPO11/RSC3-SPO11 spo11Δ/spo11Δ (AND2006), TALE-SPO11/TALE-SPO11 spo11Δ/spo11Δ (AND2529), QQR-SPO11/QQR-SPO11 spo11Δ/spo11Δ (ORD8146), SPO11/SPO11 + gRNA-YCR048W-Gal4UAS1 (ANT2524), dCAS9-SPO11/0 + gRNA-handle spo11Δ/spo11Δ (ANT2527) and dCAS9-SPO11/0 + gRNA-YCR048W-Gal4UAS1spo11Δ/spo11Δ (ANT2528) diploids, digested with AseI and probed with an internal YCR048W fragment.
Figure 3A: genomic DNA was prepared from SPO11/SPO11 GAL4/GAL4 (ORD7304), SPO11/SPO11 gal4Δ/gal4Δ (AND2599), GAL4-SPO11/GAL4-SPO11 spo11Δ/spo11Δ GAL4/GAL4 (AND2096), GAL4-SPO11/GAL4-SPO11 spo11Δ/spo11Δ gal4Δ/gal4Δ (AND1969), TALE-SPO11/TALE-SPO11 spo11Δ/spo11Δ GAL4/GAL4 (AND2529) and TALE-SPO11/TALE-SPO11 spo11Δ/spo11Δ gal4Δ/gal4Δ (AND2540) diploids, digested with XbaI and probed with a GAL2 fragment.
Figure 3B: genomic DNA was prepared from dCAS9-SPO11/0 + gRNA-handle spo11Δ/spo11Δ (ANT2527), dCAS9-SPO11/0 + gRNA-handle spo11Δ/spo11Δ gal4Δ/gal4Δ (ANT2536), dCAS9-SPO11/0 + gRNA-GAL2prom-Gal4UAS-Aspo11Δ/spo11Δ (ANT2532), dCAS9-SPO11/0 + gRNA-GAL2prom-Gal4UAS-Aspo11Δ/spo11Δ gal4Δ/gal4Δ (ANT2534), dCAS9-SPO11/0 + gRNA-GAL2prom-Gal4UAS-Bspo11Δ/spo11Δ (ANT2531), dCAS9-SPO11/0 + gRNA-GAL2prom-Gal4UAS-Bspo11Δ/spo11Δ gal4Δ/gal4Δ (ANT2535), dCAS9-SPO11/0 + gRNA-GAL2prom-Gal4UASD/Espo11Δ/spo11Δ (ANT2530), dCAS9-SPO11/0 + gRNA-GAL2prom-Gal4UAS-D/Espo11Δ/spo11Δ gal4Δ/gal4Δ (ANT2533), dCAS9-SPO11/0 + gRNAs targeting GAL2prom-Gal4UAS-A,-B,-D/Espo11Δ/spo11Δ (ANT2551) and gRNAs targeting GAL2prom-Gal4UAS-A,-B,-D/Espo11Δ/spo11Δ gal4Δ/gal4Δ (ANT2552) diploids, digested with XbaI and probed with an internal GAL2 fragment.
Figure 4A: genomic DNA was prepared from SPO11/SPO11 (ORD7304) and TEC1-SPO11/TEC1-SPO11 spo11Δ/spo11Δ (AND1926) diploids, digested with PstI and probed with an internal MSG5 fragment.
Figure 4B: genomic DNA was prepared from SPO11/SPO11 (ORD7304) and RSC3-SPO11/RSC3-SPO11 spo11Δ/spo11Δ (AND2006) diploids, digested with PstI and probed with a SIW14 fragment.
Figure 4C: genomic DNA was prepared from SPO11/SPO11 (ORD7239), QQR-SPO11/QQR-SPO11 spo11Δ/spo11Δ (RS/RS) (ORD8146), QQR-SPO11/QQR-SPO11 spo11Δ/spo11Δ (RS/rs) (ORD8683) and QQR-SPO11/QQR-SPO11 spo11Δ/spo11Δ (rs/rs) (ORD8694) diploids, digested withHincII and probed with an RSC9 fragment.
Figure 4D: genomic DNA was prepared from SPO11/SPO11 (ORD7304), dCAS9-SPO11/0 + gRNA-handle spo11Δ/spo11Δ (ANT2527) and dCAS9-SPO11/0 + gRNA-MSC1prom spo11Δ/spo11Δ (ANT2708) diploids, digested with AseI and probed with a MSC1 fragment.
Figure 6A: genomic DNA was prepared from SPO11/SPO11 (ORD7304), dCAS9-SPO11/0 spo11Δ/spo11Δ + gRNA-handle (ANT2527), GAL4-SPO11/GAL4-SPO11 spo11Δ/spo11Δ (AND2096), dCAS9-SPO11/0 spo11Δ/spo11Δ + gRNA-YCR048W-Gal4UAS1 (ANT2559) and dCAS9-SPO11/0 spo11Δ/spo11Δ + gRNA-YCR048W-Gal4UAS2 (ANT2528) diploids, digested with AseI and SacI and probed with a YCR048W-YCR051W fragment.
Figure 6B: genomic DNA was prepared from SPO11/SPO11 (ORD7304), dCAS9-SPO11/0 spo11Δ/spo11Δ + gRNA-handle (ANT2527) and dCAS9-SPO11/0 spo11Δ/spo11Δ + gRNA-SWC3 (ANT2564) diploids, digested with AvrII and PacI and probed with a MDM10-SPO7 fragment.
Figure 6C: genomic DNA was prepared from SPO11/SPO11 GAL4/GAL4 (ORD7304), dCAS9-SPO11/0 + gRNA-handle spo11Δ/spo11Δ GAL4/GAL4 (ANT2527), dCAS9-SPO11/0 + gRNA-PUT4-Gal4UASspo11Δ/spo11Δ GAL4/GAL4 (ANT2547), dCAS9-SPO11/0 + gRNA-PUT4-Gal4UASspo11Δ/spo11Δ gal4Δ/gal4Δ (ANT2553), GAL4-SPO11/GAL4-SPO11 spo11Δ/spo11Δ GAL4/GAL4 (AND2096) and GAL4-SPO11/GAL4-SPO11 spo11Δ/spo11Δ gal4Δ/gal4Δ (AND1969) diploids, digested with BamHI and XhoI and probed with a CIN1 fragment.
RESULTS
Design of targeting Spo11 fusions
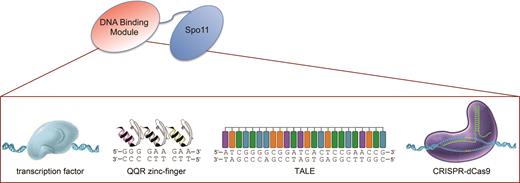
To direct Spo11 to various sites in the Saccharomyces cerevisiae genome, we fused the N-terminal region of the full length ScSPO11 gene to various DNA-binding modules and expressed the constructs under the ADH1 constitutive promoter (Figure 1 and Materials and Methods). To use the CRISPR system, we similarly integrated the PADH1-dCAS9-SPO11 construct at the TRP1 locus and expressed gRNA(s) on a high-copy 2μ plasmid (Materials and Methods). As a negative control, we used a gRNA lacking the 20-nt guide sequence (gRNA handle) but able to form the ribonucleoprotein complex (40). Unless specified, the performance of the fusion constructs was examined in diploid cells deleted for the endogenous SPO11 gene. Altogether, we generated four ‘non-programmable’ Spo11 fusions with the full-length transcription factor proteins Gal4, Tec1 and Rsc3, that recognize distinct genomic targets, or Mtw1, a subunit of the kinetochore MIND protein complex (Supplementary Table S3). In addition, we built three ‘programmable’ TSFs using an artificial three zinc-finger DNA-binding array (QQR) (41), a TALE DNA-binding array (42,43) or the Streptococus pyogenes nuclease-dead Cas9 D10A/H840A protein (dCas9) (32). QQR has 149 potential GGGGAAGAA binding sites in the yeast genome. The TALE was designed to bind a unique 25-nt target sequence (Gal4UAS-D/E) located within the promoter of the GAL2 gene. The 20-bp targets of dCas9-Spo11 were specified with various gRNAs (Supplementary Table S2).
Architecture of the targeting Spo11 fusions. To target meiotic DSB formation, the DSB-promoting Spo11 transesterase was fused to various DNA binding modules. Four different non-programmable Spo11 fusions were designed upon conjugation with the Gal4, Tec1 or Rsc3 transcription factors, or to the centromere-associated Mtw1 protein. Three programmable Spo11 fusions were constructed with an artificial zinc-finger array, a TALE polypeptide, or the CRISPR dCas9 protein. Other functional features of these DNA-binding modules are indicated in Supplementary Table S3.
The Spo11 fusions are functional
In the S. cerevisiae SK1 strain background, spore viability is very high (>90%) and is abolished in the spo11Δ mutant (Table 1). All of the SPO11 fusion constructs, expressed in one or two copies, restored >90% spore viability (Table 1). This demonstrates that the Spo11 moiety of the fusions is functional and allows the formation of a sufficient number of crossovers per chromosome to ensure their proper segregation during meiosis.
Targetable Spo11 fusions yield viable meiotic products
| . | Spore viability (%) . | |
|---|---|---|
| TSF gene construct . | One copy . | Two copies . |
| SPO11 | — | 98 (53) |
| spo11 | 93 (22) | 0 (32) |
| GAL4BD-SPO11 | ND | 96 (138)a |
| GAL4-SPO11 | 94 (18) | 92 (22) |
| TEC1-SPO11 | 88 (22) | 92 (22) |
| RSC3-SPO11 | 93 (21) | 93 (21) |
| MTW1-SPO11 | 100 (22) | 98 (24) |
| TALE-SPO11 | 97 (35) | 95 (15) |
| QQR-SPO11 | 97 (24) | 96 (24) |
| dCAS9-SPO11 | 93 (60) | 92 (22) |
| . | Spore viability (%) . | |
|---|---|---|
| TSF gene construct . | One copy . | Two copies . |
| SPO11 | — | 98 (53) |
| spo11 | 93 (22) | 0 (32) |
| GAL4BD-SPO11 | ND | 96 (138)a |
| GAL4-SPO11 | 94 (18) | 92 (22) |
| TEC1-SPO11 | 88 (22) | 92 (22) |
| RSC3-SPO11 | 93 (21) | 93 (21) |
| MTW1-SPO11 | 100 (22) | 98 (24) |
| TALE-SPO11 | 97 (35) | 95 (15) |
| QQR-SPO11 | 97 (24) | 96 (24) |
| dCAS9-SPO11 | 93 (60) | 92 (22) |
Tetrads from diploid strains were dissected and spore viability was measured (%). The number of tetrads dissected is reported in parentheses. All strains harbouring one or 2 copies of the Spo11 fusion gene construct were built in a spo11Δ/spo11Δ genetic background.
adata from (21). ND: not determined.
| . | Spore viability (%) . | |
|---|---|---|
| TSF gene construct . | One copy . | Two copies . |
| SPO11 | — | 98 (53) |
| spo11 | 93 (22) | 0 (32) |
| GAL4BD-SPO11 | ND | 96 (138)a |
| GAL4-SPO11 | 94 (18) | 92 (22) |
| TEC1-SPO11 | 88 (22) | 92 (22) |
| RSC3-SPO11 | 93 (21) | 93 (21) |
| MTW1-SPO11 | 100 (22) | 98 (24) |
| TALE-SPO11 | 97 (35) | 95 (15) |
| QQR-SPO11 | 97 (24) | 96 (24) |
| dCAS9-SPO11 | 93 (60) | 92 (22) |
| . | Spore viability (%) . | |
|---|---|---|
| TSF gene construct . | One copy . | Two copies . |
| SPO11 | — | 98 (53) |
| spo11 | 93 (22) | 0 (32) |
| GAL4BD-SPO11 | ND | 96 (138)a |
| GAL4-SPO11 | 94 (18) | 92 (22) |
| TEC1-SPO11 | 88 (22) | 92 (22) |
| RSC3-SPO11 | 93 (21) | 93 (21) |
| MTW1-SPO11 | 100 (22) | 98 (24) |
| TALE-SPO11 | 97 (35) | 95 (15) |
| QQR-SPO11 | 97 (24) | 96 (24) |
| dCAS9-SPO11 | 93 (60) | 92 (22) |
Tetrads from diploid strains were dissected and spore viability was measured (%). The number of tetrads dissected is reported in parentheses. All strains harbouring one or 2 copies of the Spo11 fusion gene construct were built in a spo11Δ/spo11Δ genetic background.
adata from (21). ND: not determined.
DSB formation at natural sites
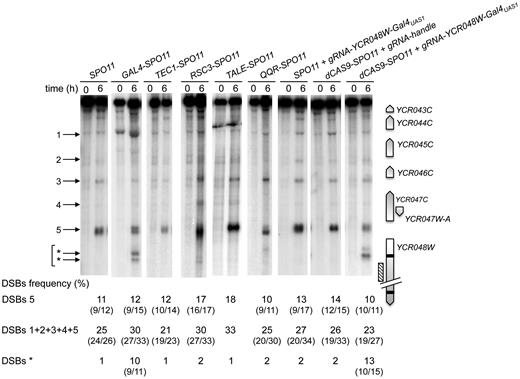
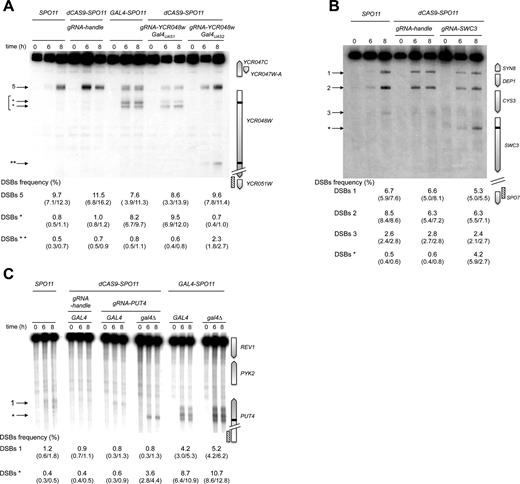
Next, we investigated whether the Spo11 fusions retain the ability to promote meiotic DSB formation in a natural DSB region. We examined the well characterized YCR043C-YCR048W region of chromosome III (44,45) that exhibits DSBs in several promoters. All TSF strains display meiotic DSBs at the same locations as in the wild-type (SPO11) strain within this 9-kb region, with the total DSB frequency varying between 21 and 33% (Figure 2). The YCR048W DSB hotspot (DSB#5) remains the strongest in all strains, ranging between 10–18% (11% in the SPO11 strain). The Gal4-Spo11 and dCas9-Spo11 (with gRNAUAS1) fusions also induce DSBs at a new site within the YCR048W coding sequence, where they have a specific binding sequence (see below).
Targeting Spo11 fusions trigger DSB formation in the natural hotspot YCR043C-YCR048W region. DSB formation was analyzed by Southern blot in cells harvested 0 and 6 h after transfer into sporulation medium. At the right of the blot, a map shows the ORFs (open arrows indicate transcriptional sense) and the position of the probe (hatched rectangle). Target GalUAS1 and GalUAS2 sites are shown as black bars. At the left of the blot, arrowheads indicate wild-type (numbered) and Gal4-Spo11 and dCas9-Spo11 targeted DSBs (asterisks). DSB frequencies (percentage of DSB fragment per total DNA) listed under each lane correspond to the mean of two independent experiments with minimum and maximum values indicated into parentheses, excepted for TALE-SPO11 (a single experiment was performed). The sum of DSB frequencies in the YCR044C (1), YCR045C (2), YCR046C (3), YCR047W-A (4), and YCR047C-YCR048W (5) promoters account for ≥ 95% of the total DSBs detected in the YCR043C-YCR048W region. Frequency of DSBs* corresponds to the sum of frequencies detected in the YCR048W ORF (indicated by *arrows).
DSB formation in cold promoter regions
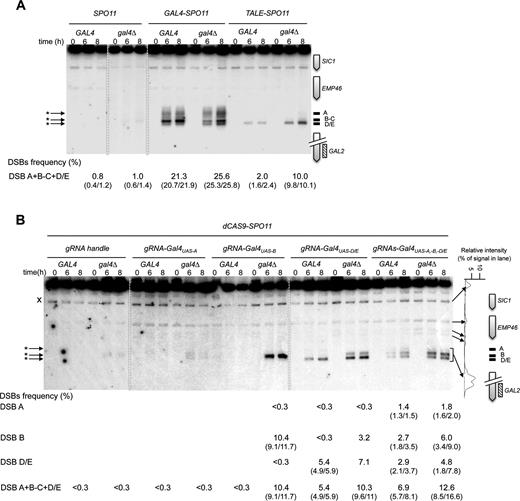
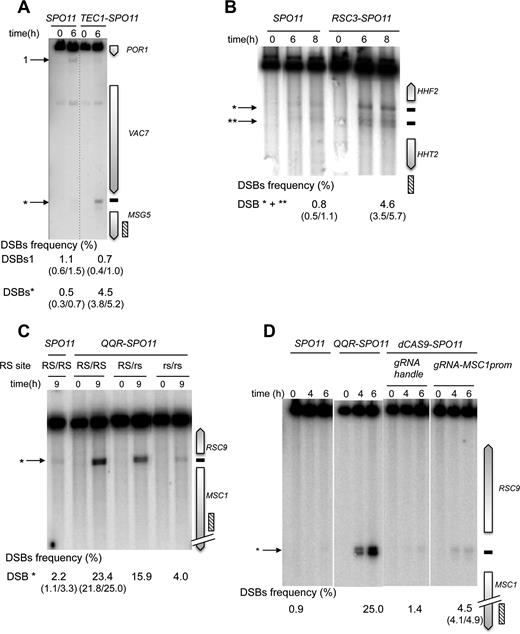
In yeasts, 90% of DSB hotspots cluster in intergenic regions containing promoters, and only ∼15% of the promoters exhibit strong hotspots (5,18). To investigate the capacity of the Spo11 fusions to mediate DSB formation in promoters that naturally exhibit low or undetectable DSBs, we examined several promoter-containing regions for each construct. Remarkably, the Gal4-, Tec1- and Rsc3-Spo11 fusions stimulated the frequency of DSB formation 10–26, 9–14 and 6–12 fold, respectively, in the targeted regions (Figures 3 and 4 and Supplementary Figure S2). Like Gal4BD-Spo11 (25,26), Gal4-Spo11 induces multiple DSBs in the promoter of the GAL2 gene that contains five partially overlapping Gal4UAS sites (Figure 3A). In the absence of the native Gal4 protein, we found that the DSB frequency was slightly enhanced from 21.3 to 25.6% (see below for detailed description of the target-binding competition of the endogenous Gal4 protein). Rsc3-Spo11 and Tec1-Spo11 precisely target two adjacent sites present in the HHF2-HHT2 and TEC1-UBC4 promoters, respectively (Figure 4B and Supplementary Figure S2).
Gal4-, TALE- and dCas9-Spo11 fusions promote DSBs in the targeted DSB-cold GAL2 promoter. Southern blot analysis of DSBs targeted by Gal4- and TALE-Spo11 (A), and dCas9-Spo11 (B) at the GAL2 promoter in GAL4 and gal4Δ cells. TSF target sites are shown as black bars at the right of the gel and arrowheads indicate TSF-induced DSBs (asterisks) at the left of the gel; crosses indicate cross hybridizing bands. The profile of DSB quantification in dCAS9-SPO11 gal4Δ diploids expressing multiple gRNAs directed against the GAL2 promoter (t = 8 h) is shown to the right of the panel B. DSB values are from single experiments or the mean of two independent experiments with minimum and maximum values indicated into parentheses.
Tec1-, Rsc3 and QQR-Spo11 fusions stimulate DSB formation in targeted DSB-cold promoter-containing regions. Southern blot analysis of DSBs targeted by Tec1-Spo11 at the MSG5 promoter (A), Rsc3-Spo11 at the HHF2-HHT2 promoter region (B) and QQR-Spo11 and dCas9-Spo11 at the RSC9-MSC1 promoters (C and D). TSF target sites are shown as black bars at the right of the gel and arrowheads indicate wild-type (numbered) and TSF-induced DSBs (asterisks) at the left of the gel. The presence of the homozygous wild-type QQR recognition site (RS), heterozygous mutated RS (RS/rs) and homozygous mutated RS (rs/rs) is indicated in panel C. DSB values as described in Figure 3 legends.
Regarding the programmable Spo11 fusions, QQR-Spo11 also strongly stimulates DSB formation in regions that contain a recognition site. Namely, it increases 10-fold (2.2–23.4%) within the RSC9-MSC1 promoter region (Figure 4C) and 4-fold (2.0 versus 8.0%) in the ESI1-HOF1 promoter region, where it overlaps a natural DSB site (Supplementary Figure S3A). In the SUT1-RAD54 promoter region QQR-Spo11 triggers additional DSBs in the vicinity of the target site (Supplementary Figure S3B); the DSB#1 is located near a 1 bp variant QQR site (GGGGAAGAC). The other DSBs are weaker and unexpected. It can result from a local positive cis-effect increasing chromatin accessibility near the target site or from the local interaction of QQR with unidentified chromatin/DNA associated proteins. To test QQR target specificity, we mutated two triplets of the QQR recognition site (RS) located in the RSC9-MSC1 region. In the strain heterozygous for the target site (RS/rs), DSB formation was reduced from 23.4 to 15.9% and was further reduced near the wild type level (4.0%) in the rs/rs strain mutated on both homologs (Figure 4C), demonstrating that QQR-Spo11 recognizes the expected target sequence in vivo. Concerning targeting Spo11 with a TALE module, DSB frequency only slightly increases (0.8 versus 2.0%) at the Gal4UAS-D/E target site located within the GAL2 promoter (Figure 3A), due to endogenous binding competition (see below). DSB formation was abolished upon mutation of its binding sequence (Supplementary Figure S4A). Based on these results, we concluded that the QQR and TALE programmable modules also allow the functional targeting of Spo11.
Then, to test dCas9-Spo11 and compare its efficiency to QQR-, Gal4- and TALE-Spo11 DSB formation in the same region, we designed several gRNAs matching the QQR and Gal4 binding sites in the RSC9-MSC1 and GAL2 promoter regions, respectively. Directing dCas9-Spo11 to the QQR binding site led to a 3-fold increase of DSB frequency at the target site (1.4 versus 4.5%; Figure 4D). In the GAL2 promoter, co-expression of dCas9-Spo11 with a gRNA-Gal4UAS-D/E led to a 18-fold stimulation of targeted DSBs (5.4%) compared to the controls (<0.3%), but surprisingly, targeting of the adjacent Gal4UAS-A and Gal4UAS-B sites was not efficient (Figure 3B). However, multiplex targeting of the Gal4UAS-A, -B, -D/E sites resulted in an enhanced DSB frequency (6.9%) distributed in three discrete bands, whose migration corresponds to DSB formation at each target site (Figure 3B). For comparison, the total DSB frequency in the GAL2 promoter appears the highest in the GAL4-SPO11 strain (21.3%) with five target Gal4UAS binding sites, intermediate in the dCAS9-SPO11 strain (up to 6.9% upon multiplex targeting) and weak in the TALE-SPO11 strain (2.0%) targeting the single Gal4UAS-D/E site.
Altogether these results demonstrate that distinct DNA binding modules, i.e. full length transcription factors, synthetic Zn fingers, TALE domains and dCas9, can be conjugated to Spo11 in order to induce meiotic DSB formation in various DSB-cold promoters harbouring a binding site, but local- and module-specific variations in efficiency are observed.
Competition with endogenous factors modulates the targeting efficiency
One potential limitation to targeting DSBs with Spo11 fusions is the local competition with endogenous DNA- or chromatin-binding protein(s). In vitro studies have shown that the Gal4 protein displays nanomolar affinity for the consensus Gal4UAS binding sequences (46,47). To investigate whether the native Gal4 protein interferes with targeting the Gal4UAS, we expressed the Gal4-, TALE- and dCas9-Spo11 fusions in strains deleted for the endogenous GAL4 gene. As a control, deletion of GAL4 in the SPO11 strain had no stimulating effect (Figure 3A). In contrast, the Gal4-Spo11 DSB frequency was slightly enhanced at the targeted GAL2 promoter (from 21.3 to 25.6%) as previously observed for Gal4BD-Spo11 (48). Similarly, DSBs generated by Gal4-Spo11 at the GAL7 and GAL1-GAL10 promoters was increased 2.8- and 4.4-fold, respectively, in the gal4Δ strain (Supplementary Figure S2A). The targeting of DSBs at the GAL2 Gal4UAS-D/E site by TALE-Spo11 was enhanced 5-fold (up to 10%) in the absence of the native Gal4 protein (Figure 3A and Supplementary Figure S4A). As expected, TALE-Spo11 DSBs were barely (1.5%) or not detectable in the GAL2-abcdE strain mutated for the TALE binding sequence and at the other Gal4UAS sites located in the GAL1/GAL10 and GAL7 promoters (Supplementary Figure S4A and B). As a positive control, Gal4-Spo11 strongly targets DSBs in the GAL2-abcdE strain which retains only the Gal4UAS-E binding site. These results demonstrate the target specificity of the TALE array in recognizing a unique 24-bp sequence. Similarly, DSBs induced by dCas9-Spo11, using single or multiple gRNAs targeting the GAL2 Gal4UAS sites, were enhanced up to 40-fold (<0.3% versus 12.6%) in the gal4Δ mutant (Figure 3B). These results suggest that the native Gal4 protein acts as a strong competitor for the binding of the relevant Gal4-, TALE- and dCas9-Spo11 fusions.
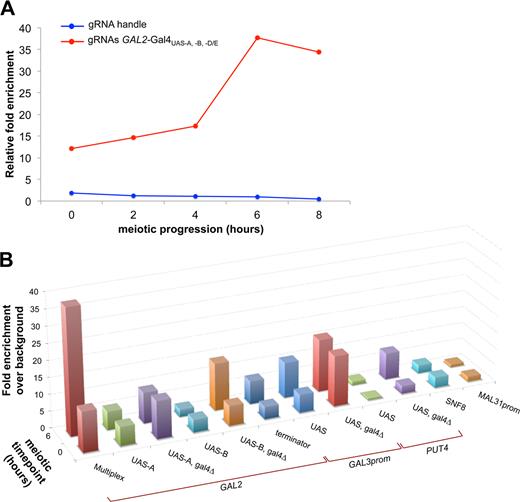
Finally, to assess whether the endogenous competition by Gal4 results from reduced binding of the Spo11 fusion proteins to the target sites, we performed ChIP-qPCR assays of the dCas9-Spo11–3xFLAG construct using an anti-FLAG antibody (Materials and Methods). In the most efficient DSB-targeting situation, i.e. dCas9-Spo11 expressed with multiple gRNAs, we readily detected chromatin binding of dCas9-Spo11 at the GAL2 locus at the onset of meiosis (t = 0 h) and observed the expected increase in association during the prophase of meiosis, reaching a 3-fold higher level at t = 6 h, when DSB formation occurs (Figure 5A). The gRNA handle control showed no dCas9-Spo11 binding. The binding of dCas9-Spo11 upon co-expression of the gRNA-Gal4UAS-A and gRNA-Gal4UAS-B was low and increased 2- and 7-fold upon removal of the Gal4 protein (Figure 5B), respectively. Mechanistically, these results indicate that the binding of CRISPR-dCas9-Spo11 to the target site is not sufficient for cleavage (Gal4UAS-A) and is sensitive to the competition with an endogenous protein (Gal4UAS-B).
dCas9-Spo11 fusion binding at various genomic loci. Quantitative analysis of dCas9-Spo11 association with the target GAL2 promoter upon expression of the gRNA handle or multiple gRNAs (A), and with various genomic loci upon expression of the corresponding gRNAs (B), was assessed by Chromatin immunoprecipitation (ChIP) in sae2Δ diploids expressing the Flag-tagged dCas9-Spo11 protein. Cells were collected at the indicated times after transfer to sporulation medium (t = 0 h), fixed with formaldehyde in vivo and immunoprecipitated with anti-flag antibodies. Quantitation of immunoprecipitated DNA was performed via qPCR and fold enrichments are expressed as the relative fold enrichment of target site over SMC1 signal and are normalized to input DNA (Materials and Methods). In the panel B the enrichment for each target site was normalized to the fold enrichment measured in control gRNA handle-dCAS9-SPO11 cells.
DSB induction in coding regions
In yeasts, <5% of the DSB hotspots are located in non-promoter regions (5,18). To assay the performance of the various targeting strategies, we first examined DSB formation in the YCR048W coding region that contains two Gal4 consensus binding sequences, Gal4UAS1 and Gal4UAS2, located at the beginning and the end of the coding region, respectively. In SPO11 diploids, rare DSBs (1%) are detected near the YCR048W-Gal4UAS1 (Figure 2). However, like Gal4BD-Spo11 (21), Gal4-Spo11 and dCas9-Spo11 stimulate DSB formation at the Gal4UAS1 site (up to 13-fold) (Figure 2). As controls, co-expression of dCas9-Spo11 and a gRNA handle in spo11Δ cells, as well as the sole expression of the gRNA-YCR048W-Gal4UAS1 in SPO11 cells failed to stimulate DSB formation in this coding region. Concerning the downstream Gal4UAS2, neither Gal4BD-Spo11 nor Gal4-Spo11 stimulates DSBs at this site [(21) and Figure 6A]. However, cleavage is slighltly enhanced in cells co-expressing dCas9-Spo11 and the gRNA-YCR048W-Gal4UAS2, reaching 2.3%.
Spo11 fusion DSB formation in targeted gene coding sequences. Southern blot analysis of Gal4-Spo11 and dCas9-Spo11 DSB formation at the target YCR048W (A), SWC3 (B) and PUT4 (C) ORFs. Gal4-Spo11 and dCas9-Spo11 target sites are shown as black bars at the right of the gel and arrowheads indicate wild-type (numbered) and targeted DSBs (asterisks) at the left of the gel. DSB values as described in Figure 3 legends.
Next, we examined the targeting efficiency of dCas9-Spo11-mediated DSBs in the cold SWC3 coding region located a gene away from the natural CYS3 and DEP1-SYN8 hot spots (49). Remarkably, the strain expressing the dCas9-Spo11 and gRNA-SWC3 strongly induces DSB formation in the targeted region (0.6% versus 4.2% ), while retaining DSBs in the CYS3 and DEP1 promoters as in wild type (Figure 6B). We also designed gRNAs directed against the Gal4UAS sites contained in the GAL2, PUT4, NCE102 and YPR1 genes located in naturally cold regions (26). DSB formation in the PUT4 region is weak in the wild-type strain (1.6%) but is strongly stimulated by Gal4-Spo11 in the presence or absence of the Gal4 protein, reaching 12.9 and 15.9%, respectively (Figure 6C). In contrast, removal of Gal4 was required to stimulate DSB formation by dCas9-Spo11, reaching 4.4%. This dependency is consistent with the high level of dCas9-Spo11 chromatin binding to the PUT4-Gal4UAS target (8-fold enrichment over background), only in the absence of Gal4 (Figure 5B). No Gal4-Spo11 nor dCas9-Spo11 targeted DSBs were observed in the coding region of the GAL2, NCE102 and YPR1 genes, in the presence or absence of Gal4 (Supplementary Figure S5). Altogether, these results demonstrate that DSB formation can be stimulated in coding regions, but the performance is heterogeneous.
Targeting of DSB refractory regions
Next, we examined the targeting of dCas9-Spo11 in other known DSB-rare regions. No targeted DSB formation was observed in the terminator regions of the SWC3 and GAL2 genes (Supplementary Figure S6), albeit dCas9-Spo11 associates with the latter locus (Figure 5B). We also attempted to stimulate DSB formation in pericentric regions that are universally cold for DSB formation (5,7,14,18,26,50,51). Neither natural nor targeted DSBs were observed in the SNF8 gene located 1.8 kb from CEN16 (Supplementary Figure S7A), in agreement with the lack of chromatin binding (Figure 5B). Whereas dCas9-Spo11 binds to the Gal4UAS-containing GAL3 region (located 13 kb from the centromere of chromosome IV) in a Gal4p-independent manner (Figure 5B), no natural nor targeted DSB formation was detected (Supplementary Figure S7D). We also built a fusion of Spo11 to the centromere-associated Mtw1 protein, a non-essential component of the kinetochore complex (52) (Supplementary Table S3). Albeit expression of Mtw1-Spo11 restored spore viability in the absence of Spo11 (Table 1), no DSB formation was detected near the centromere of chromosomes XVI and III (Supplementary Figure S7B and C). These results confirm the strong inhibitory effects on DSB formation exerted by a centromere (26,50,53,54).
Subtelomere regions are cold in yeast but rather recombination proficient in other eukaryotes, including plants and mammals (17,55). We designed a gRNA to direct dCas9-Spo11 to a unique subtelomeric sequence located ∼10 kb from the end of chromosome II. Neither chromatin binding nor DSB formation was detected in the targeted MAL31 promoter region (Figure 5B and Supplementary Figure S8). Finally, to attempt the targeting of a repeated sequence of the yeast genome, we assayed the targeting of Spo11 to the ∼330-bp long terminal repeat (LTR) sequences which flank the transposable Ty elements (30 copies per SK1 cell (56)). The Tys are located in various naturally DSB-prone or -cold regions of the genome, nevertheless with the common feature that they are internally cold but not fully refractory (56). We programmed a single gRNA to match a unique 20-nt sequence conserved in the LTR sequence of 11 Tys (56) and examined DSB formation in two different genomic contexts. In the TyPEX25-CAR1 region natural DSB formation was slightly enhanced near the 5′ LTR (15.8 versus 25.3%) but no DSBs were detected in both targeted 5′ and 3′ LTRs per se (Supplementary Figure S9A-B). In contrast, in the naturally cold TyCUS2-MRP10 region, we observed a 3-fold increase (0.4 versus 1.3%) in DSB formation within the 5′ LTR (Supplementary Figure S9C). No DSB formation was detected in the 3′ LTR (Supplementary Figure S9D).
These results show that the targeting of DSBs remains inefficient in certain regions of the genome even with dCas9-Spo11, indicating that unlocking DSB formation in these refractory regions will require other strategies.
DSB targeting locally enhances crossover frequency
To verify that the targeted stimulation of DSB formation enhances meiotic recombination frequencies, we introduced flanking heterozygous markers in several chromosomal regions and measured the meiotic recombination rates in the progenies of each TSF strain, upon dissection of four-spore tetrads. In all cases, except negative controls, we observed a significant 2- to 6-fold stimulation of crossovers (calculated as cM/kb). This was true for the Spo11 fusions with Gal4, Tec1, Rsc3, QQR, TALE and dCas9 (Table 2) at sites where DSB formation was observed. For comparison, in the GAL2 region, Gal4BD- and Gal4-Spo11 fusions resulted in a 5.4- and 5.0-fold stimulation of crossovers compared to the wild type SPO11 strain while TALE-Spo11 (in the most stimulating gal4Δ context) and the single and multiplexed targeting of dCas9-Spo11 induced a 3.1-, 2.7- and 6.3-fold increase of crossover frequencies, respectively. The negative controls (expression of the gRNA handle or a gRNA directed against the non-matching SWC3 gene sequence) show no crossover stimulation above wild type. To note, as DSBs, the crossover frequency was also stimulated at the PUT4 region by Gal4- and dCas9-Spo11 (3- and 4-fold, respectively) upon deletion of the GAL4 gene. In the MSC1 region targeted by QQR-Spo11, recombination is enhanced 5.6-fold over the background and is dependent on the presence of the RS motif. In contrast, no significant stimulation of recombination was observed with the targeting of dCas9-Spo11, consistent with the low increase of targeted DSBs compared to QQR-Spo11 DSBs. Taken together, these results show that like Gal4BD-Spo11 (21,25), the targeting of Spo11 with different types of DNA binding modules increases the meiotic recombination frequency.
The Spo11 fusions stimulate meiotic recombination in the targeted regions
| Genotype . | Locus . | Target site(s) . | Targeted DSB frequency (%) . | Recombination rate (cM/kb) . | Stimulation versus WT . |
|---|---|---|---|---|---|
| SPO11/'' | GAL2 | — | 0.8 | 0.5 | — |
| GAL4BD-SPO11/'' a | GAL2 | ABCD/E | 21.1 | 2.6 | 5.4 |
| GAL4-SPO11/'' | GAL2 | ABCD/E | 21.3 | 2.5 | 5.0 |
| TALE-SPO11 /‘' gal4Δ/'’ | GAL2 | D/E | 10.0 | 1.5 | 3.1 |
| dCAS9-SPO11/- +gRNA handle | GAL2 | — | < 0.3 | 0.6 | 1.2* |
| dCAS9-SPO11/- +gRNA-SWC3 | GAL2 | — | < 0.3 | 0.5 | 1.0* |
| dCAS9-SPO11/- +gRNA-UAS-D/E | GAL2 | D/E | 5.4 | 1.3 | 2.7 |
| dCAS9-SPO11/- + gRNAs-UAS-A, -B, -D/E | GAL2 | ABD/E | 6.9 | 3.1 | 6.3 |
| SPO11/” | PUT4 | — | 1.6 | 0.9 | — |
| GAL4-SPO11/'' | PUT4 | RS | 12.9 | 2.2 | 2.3 |
| SPO11/" gal4Δ/'' | PUT4 | RS | 1.6 | 0.9 | 1.0* |
| GAL4-SPO11/‘' gal4Δ/'’ | PUT4 | RS | 15.9 | 2.9 | 3.2 |
| dCAS9-SPO11/- + gRNA handle gal4Δ/'' | PUT4 | RS | 1.3 | 1.8 | 1.9* |
| dCAS9-SPO11/- + gRNA-UASgal4Δ/'' | PUT4 | RS | 4.4 | 3.7 | 4.0 |
| SPO11/” | MSG5 | — | 1.6 | 0.1 | — |
| TEC1-SPO11/'' | MSG5 | RS | 5.2 | 0.6 | 4.6 |
| SPO11/” | HHF2 | — | 0.8 | 0.1 | — |
| RSC3-SPO11/'' | HHF2 | RS | 4.6 | 0.6 | 6.3 |
| SPO11/'' | MSC1 | — | 2.2 | 0.3 | — |
| QQR-SPO11/'' | MSC1 | RS | 23.4 | 1.7 | 5.6 |
| QQR-SPO11/'' | MSC1 | rs | 4.0 | 0.2 | 0.6* |
| dCAS9-SPO11/- +gRNA handle | MSC1 | RS | 1.4 | 0.2 | 0.7* |
| dCAS9-SPO11/- +gRNA-MSC1prom | MSC1 | RS | 4.5 | 0.3 | 1.0* |
| Genotype . | Locus . | Target site(s) . | Targeted DSB frequency (%) . | Recombination rate (cM/kb) . | Stimulation versus WT . |
|---|---|---|---|---|---|
| SPO11/'' | GAL2 | — | 0.8 | 0.5 | — |
| GAL4BD-SPO11/'' a | GAL2 | ABCD/E | 21.1 | 2.6 | 5.4 |
| GAL4-SPO11/'' | GAL2 | ABCD/E | 21.3 | 2.5 | 5.0 |
| TALE-SPO11 /‘' gal4Δ/'’ | GAL2 | D/E | 10.0 | 1.5 | 3.1 |
| dCAS9-SPO11/- +gRNA handle | GAL2 | — | < 0.3 | 0.6 | 1.2* |
| dCAS9-SPO11/- +gRNA-SWC3 | GAL2 | — | < 0.3 | 0.5 | 1.0* |
| dCAS9-SPO11/- +gRNA-UAS-D/E | GAL2 | D/E | 5.4 | 1.3 | 2.7 |
| dCAS9-SPO11/- + gRNAs-UAS-A, -B, -D/E | GAL2 | ABD/E | 6.9 | 3.1 | 6.3 |
| SPO11/” | PUT4 | — | 1.6 | 0.9 | — |
| GAL4-SPO11/'' | PUT4 | RS | 12.9 | 2.2 | 2.3 |
| SPO11/" gal4Δ/'' | PUT4 | RS | 1.6 | 0.9 | 1.0* |
| GAL4-SPO11/‘' gal4Δ/'’ | PUT4 | RS | 15.9 | 2.9 | 3.2 |
| dCAS9-SPO11/- + gRNA handle gal4Δ/'' | PUT4 | RS | 1.3 | 1.8 | 1.9* |
| dCAS9-SPO11/- + gRNA-UASgal4Δ/'' | PUT4 | RS | 4.4 | 3.7 | 4.0 |
| SPO11/” | MSG5 | — | 1.6 | 0.1 | — |
| TEC1-SPO11/'' | MSG5 | RS | 5.2 | 0.6 | 4.6 |
| SPO11/” | HHF2 | — | 0.8 | 0.1 | — |
| RSC3-SPO11/'' | HHF2 | RS | 4.6 | 0.6 | 6.3 |
| SPO11/'' | MSC1 | — | 2.2 | 0.3 | — |
| QQR-SPO11/'' | MSC1 | RS | 23.4 | 1.7 | 5.6 |
| QQR-SPO11/'' | MSC1 | rs | 4.0 | 0.2 | 0.6* |
| dCAS9-SPO11/- +gRNA handle | MSC1 | RS | 1.4 | 0.2 | 0.7* |
| dCAS9-SPO11/- +gRNA-MSC1prom | MSC1 | RS | 4.5 | 0.3 | 1.0* |
Recombination rates were measured by introduction of heterozygous genetic markers on either side of the targeted regions and their segregation examined in meiotic products upon tetrad dissection. According to the phenotype of drug resistance, the numbers of parental ditype (PD), tetratype (T) and non-parental ditype (NPD) were determined to calculate the genetic distance in centimorgans (cM) (see Supplementary Table S4 for the complete data). The difference in recombination frequency between the wild-type and TSF strains is statistically different (P-values < 0.05; two-tailed Fisher's exact test), except for those indicated by an asterisk (P-values > 0.2; two-tailed Fisher's exact test).
aData from (25).
| Genotype . | Locus . | Target site(s) . | Targeted DSB frequency (%) . | Recombination rate (cM/kb) . | Stimulation versus WT . |
|---|---|---|---|---|---|
| SPO11/'' | GAL2 | — | 0.8 | 0.5 | — |
| GAL4BD-SPO11/'' a | GAL2 | ABCD/E | 21.1 | 2.6 | 5.4 |
| GAL4-SPO11/'' | GAL2 | ABCD/E | 21.3 | 2.5 | 5.0 |
| TALE-SPO11 /‘' gal4Δ/'’ | GAL2 | D/E | 10.0 | 1.5 | 3.1 |
| dCAS9-SPO11/- +gRNA handle | GAL2 | — | < 0.3 | 0.6 | 1.2* |
| dCAS9-SPO11/- +gRNA-SWC3 | GAL2 | — | < 0.3 | 0.5 | 1.0* |
| dCAS9-SPO11/- +gRNA-UAS-D/E | GAL2 | D/E | 5.4 | 1.3 | 2.7 |
| dCAS9-SPO11/- + gRNAs-UAS-A, -B, -D/E | GAL2 | ABD/E | 6.9 | 3.1 | 6.3 |
| SPO11/” | PUT4 | — | 1.6 | 0.9 | — |
| GAL4-SPO11/'' | PUT4 | RS | 12.9 | 2.2 | 2.3 |
| SPO11/" gal4Δ/'' | PUT4 | RS | 1.6 | 0.9 | 1.0* |
| GAL4-SPO11/‘' gal4Δ/'’ | PUT4 | RS | 15.9 | 2.9 | 3.2 |
| dCAS9-SPO11/- + gRNA handle gal4Δ/'' | PUT4 | RS | 1.3 | 1.8 | 1.9* |
| dCAS9-SPO11/- + gRNA-UASgal4Δ/'' | PUT4 | RS | 4.4 | 3.7 | 4.0 |
| SPO11/” | MSG5 | — | 1.6 | 0.1 | — |
| TEC1-SPO11/'' | MSG5 | RS | 5.2 | 0.6 | 4.6 |
| SPO11/” | HHF2 | — | 0.8 | 0.1 | — |
| RSC3-SPO11/'' | HHF2 | RS | 4.6 | 0.6 | 6.3 |
| SPO11/'' | MSC1 | — | 2.2 | 0.3 | — |
| QQR-SPO11/'' | MSC1 | RS | 23.4 | 1.7 | 5.6 |
| QQR-SPO11/'' | MSC1 | rs | 4.0 | 0.2 | 0.6* |
| dCAS9-SPO11/- +gRNA handle | MSC1 | RS | 1.4 | 0.2 | 0.7* |
| dCAS9-SPO11/- +gRNA-MSC1prom | MSC1 | RS | 4.5 | 0.3 | 1.0* |
| Genotype . | Locus . | Target site(s) . | Targeted DSB frequency (%) . | Recombination rate (cM/kb) . | Stimulation versus WT . |
|---|---|---|---|---|---|
| SPO11/'' | GAL2 | — | 0.8 | 0.5 | — |
| GAL4BD-SPO11/'' a | GAL2 | ABCD/E | 21.1 | 2.6 | 5.4 |
| GAL4-SPO11/'' | GAL2 | ABCD/E | 21.3 | 2.5 | 5.0 |
| TALE-SPO11 /‘' gal4Δ/'’ | GAL2 | D/E | 10.0 | 1.5 | 3.1 |
| dCAS9-SPO11/- +gRNA handle | GAL2 | — | < 0.3 | 0.6 | 1.2* |
| dCAS9-SPO11/- +gRNA-SWC3 | GAL2 | — | < 0.3 | 0.5 | 1.0* |
| dCAS9-SPO11/- +gRNA-UAS-D/E | GAL2 | D/E | 5.4 | 1.3 | 2.7 |
| dCAS9-SPO11/- + gRNAs-UAS-A, -B, -D/E | GAL2 | ABD/E | 6.9 | 3.1 | 6.3 |
| SPO11/” | PUT4 | — | 1.6 | 0.9 | — |
| GAL4-SPO11/'' | PUT4 | RS | 12.9 | 2.2 | 2.3 |
| SPO11/" gal4Δ/'' | PUT4 | RS | 1.6 | 0.9 | 1.0* |
| GAL4-SPO11/‘' gal4Δ/'’ | PUT4 | RS | 15.9 | 2.9 | 3.2 |
| dCAS9-SPO11/- + gRNA handle gal4Δ/'' | PUT4 | RS | 1.3 | 1.8 | 1.9* |
| dCAS9-SPO11/- + gRNA-UASgal4Δ/'' | PUT4 | RS | 4.4 | 3.7 | 4.0 |
| SPO11/” | MSG5 | — | 1.6 | 0.1 | — |
| TEC1-SPO11/'' | MSG5 | RS | 5.2 | 0.6 | 4.6 |
| SPO11/” | HHF2 | — | 0.8 | 0.1 | — |
| RSC3-SPO11/'' | HHF2 | RS | 4.6 | 0.6 | 6.3 |
| SPO11/'' | MSC1 | — | 2.2 | 0.3 | — |
| QQR-SPO11/'' | MSC1 | RS | 23.4 | 1.7 | 5.6 |
| QQR-SPO11/'' | MSC1 | rs | 4.0 | 0.2 | 0.6* |
| dCAS9-SPO11/- +gRNA handle | MSC1 | RS | 1.4 | 0.2 | 0.7* |
| dCAS9-SPO11/- +gRNA-MSC1prom | MSC1 | RS | 4.5 | 0.3 | 1.0* |
Recombination rates were measured by introduction of heterozygous genetic markers on either side of the targeted regions and their segregation examined in meiotic products upon tetrad dissection. According to the phenotype of drug resistance, the numbers of parental ditype (PD), tetratype (T) and non-parental ditype (NPD) were determined to calculate the genetic distance in centimorgans (cM) (see Supplementary Table S4 for the complete data). The difference in recombination frequency between the wild-type and TSF strains is statistically different (P-values < 0.05; two-tailed Fisher's exact test), except for those indicated by an asterisk (P-values > 0.2; two-tailed Fisher's exact test).
aData from (25).
DISCUSSION
The data reported here demonstrate that increases in meiotic double-strand breaks and crossover frequencies can be programmed to arbitrary genomic sites in yeast, by fusion of the Spo11 protein to various DNA-recognition domains. These domains include full-length natural transcription factors, synthetic arrays of zinc finger or TALE modules, and the dCas9 protein in conjunction with designed gRNAs. Importantly, all these Spo11 fusions retain the capacity to form DSBs at the natural sites, thereby ensuring wild-type spore viability, even in the absence of the wild-type SPO11 gene. At the same time, inactivation of SPO11 is not necessary for the effect to be manifested (Supplementary Figure S10).
The degree of stimulation of DSB frequencies by the Spo11 fusions was quite variable, depending on the constructs and target sites. Consistently, in most of the cases the crossover frequencies at the successfully targeted loci increased (2.3- to 6.3-fold), with no adverse effect on spore viability (Table 1). In the case of dCas9-Spo11, some of the variability may be due to the quality of the gRNAs, which was not independently tested. Altogether, using Spo11 fusions, we assayed 34 target sites located in 26 different regions (summarized in Table S5) and found that the targeting efficiency varies from site to site, differs according to the TSFs and is modulated by endogenous competition and meiosis-specific binding to the chromatin (Figure 5). Hierarchically, DSB-cold promoter-containing regions are the most efficiently TSF-targeted regions (15/16 cases), suggesting that these ‘permissive’ regions are prone to be warmed-up by the Spo11 fusions. Like the natural DSB hotspots, these TSF-targeted DSBs had a low level of nucleosome occupancy (5), consistent with an open chromatin structure which promotes DNA target accessibility to Spo11 (57–59). Targeting of ORF regions was less successful (6/12 cases) and was located in the proximity of a weak (PUT4) or strong DSB hotspot (YCR048W and SWC3) region. The regional overlap with natural DSB sites likely provides favourable chromatin structure for Spo11 accessibility and cleavage. However, and as previously noted for natural hotspots (5), low nucleosome occupancy is not sufficient to warm up a cold region since TSF-targeting of GAL2 and SWC3 gene terminators was inefficient despite a low level of nucleosome occupancy. In agreement with previous results (26), TSFs inefficiently target regions near the centromeres and telomeres where natural DSBs are suppressed, perhaps to ensure proper homolog and sister chromatin segregation. Along this line, our data also show that in many cases, including pericentric regions, TSF binding to target site is not sufficient to trigger DSB formation, indicating that other essential cis and/or trans factor(s) locally regulate cleavage by Spo11 (5,26). For instance, it has been shown that the Cft19 complex inhibits DSB formation near the centromere (54). Thus so far, some genomic sites still remain refractory to DSB targeting and their warming will require further innovations.
Another factor that influences the ability to program DSBs with Spo11 fusions is occupancy of the target by endogenous DNA-binding proteins. The level of stimulation at natural Gal4 binding sites by Gal4-, TALE- and dCas9-Spo11 was enhanced by deletion of the endogenous GAL4 gene. This effect was less pronounced for Gal4-Spo11, suggesting either that it has a higher inherent affinity for its target sequence, or that it is better able to compete with the endogenous protein. This advantage may apply to other natural transcription factors as well; however, not all apparent binding sites for these proteins can be accessed. For example, only about 25 of the identified 496 Gal4UAS sites in the yeast SK1 genome are bound in vivo (26,60).
Since all TSFs were expressed under the same promoter and since the amount of TSF activity does not seem to be limiting (Table 1), a comparison of targeted recombination efficiency between the various Spo11 fusions is possible. This is summarized in Table 2. In the targeted GAL2 promoter and PUT4 gene, Gal4- and CRISPR-dCas9-Spo11 constructs induce similar level of recombination stimulation (5.0 versus 6.3 and 3.2 versus 4.0-fold, respectively and not statistically different). The fact that Gal4 binds to > 25 targets genome-wide (26,60) instead of up to three for CRISPR-dCas9 and that CRISPR-dCas9 is much more sensitive to native Gal4 target-binding competition, suggests that Gal4 has a stronger affinity for the Gal4UAS sites in vivo than the CRISPR-dCas9 system. Similarly, TALE- and CRISPR-dCas9-Spo11 lead to an approximately 3-fold enhancement of recombination frequency in the GAL2 region containing the unique target Gal4UASD/E site but TALE-Spo11 efficiency is strongly impaired by endogenous Gal4 interference. It suggests that CRISPR-dCas9 displays a higher affinity for the Gal4UASD/E site than TALE. In contrast, CRISPR-dCas9-Spo11 poorly stimulates DSBs at the MSC1 promoter while QQR-Spo11 is very proficient and enhanced approximately 6-fold the targeted crossover frequency. It suggests that QQR has a tighter binding capacity to this target site than CRISPR-dCas9-Spo11, being driven by a proficient DNA binding affinity and/or being assisted by other protein-protein chromatin interactions. Thus, choosing endogenous transcription factors from the host organism may sometimes be more efficient as transcription factors are likely evolutionarily selected proteins most able to find their target and overcome competition with other native factors. Although there are little comparative data, zinc fingers and TALE nucleases seem to be just as effective (and sometimes more effective) for particular target site than the CRISPR system, indicating again that the efficiency of the programmable Spo11 fusions can vary greatly among targets.
The disadvantage to natural factors is that they do not allow programming to arbitrary regions of the genome. In contrast, zinc fingers, TALE domains and the CRISPR-Cas9 platform can all be directed to a wide range of sequences. It is worth noting that the different programmable Spo11 fusions generate DSBs with varying degrees of efficiency at the same target locus (Figures 3 and 4D and Supplementary Figure S3B). However, dCas9-Spo11 has several advantages: design for new targets is very simple (just production of new gRNAs) and a desired target region can be investigated by testing a number of different gRNAs. Multiple gRNAs can be introduced together, and this can improve DSB formation above any single gRNA (see Figure 3B).
An alternative approach to stimulating recombination frequencies is to manipulate the processing of meiotic DSBs at the repair stages. Previous studies in A. thaliana demonstrated that stimulation of meiotic crossovers is obtained in mutant lines that alter the outcome of a subset of DSB repair events (27–30). In this case, elevated rates were only observed at natural crossover sites, along the non-interfering CO pathway. In contrast, the Spo11 fusions described here allow to modify recombination initiation and produce experimental alteration of the location and frequencies of DSBs, with the consequence to locally enhance gene conversion and crossover frequencies (21,25) (present data).
Modifying the natural meiotic recombination landscape is a highly desirable biotechnological advance. Overcoming the generally low level of recombination events per chromosome per generation (1 or 2) observed in most eukaryotes will facilitate the separation or re-association of closely-linked alleles in hybrid strains. This promises to significantly reduce the cost and timeframe for the development, for example, of novel plant varieties.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to all members of our laboratory, especially to L. Carrière and S. Loeillet for many fruitful discussions.
FUNDING
ANR [BLAN06-3-150811]; ARC and Meiogenix©; CNRS and the Institut Curie (to N.U.). Funding for open access charge: Institut Curie.
Conflict of interest statement. A.N. and G.B. are CSO and CEO, cofounders of Meiogenix, respectively. G.B. and A.S. are employed by Meiogenix. G.B., A.N. and A.S. are inventors of a patent issued from this research.
Present address: Norio Uematsu, Shiodome Building, 1-2-20 Kalgan, Minato-Ku, Tokyo 105-0022, Japan.
REFERENCES




Comments