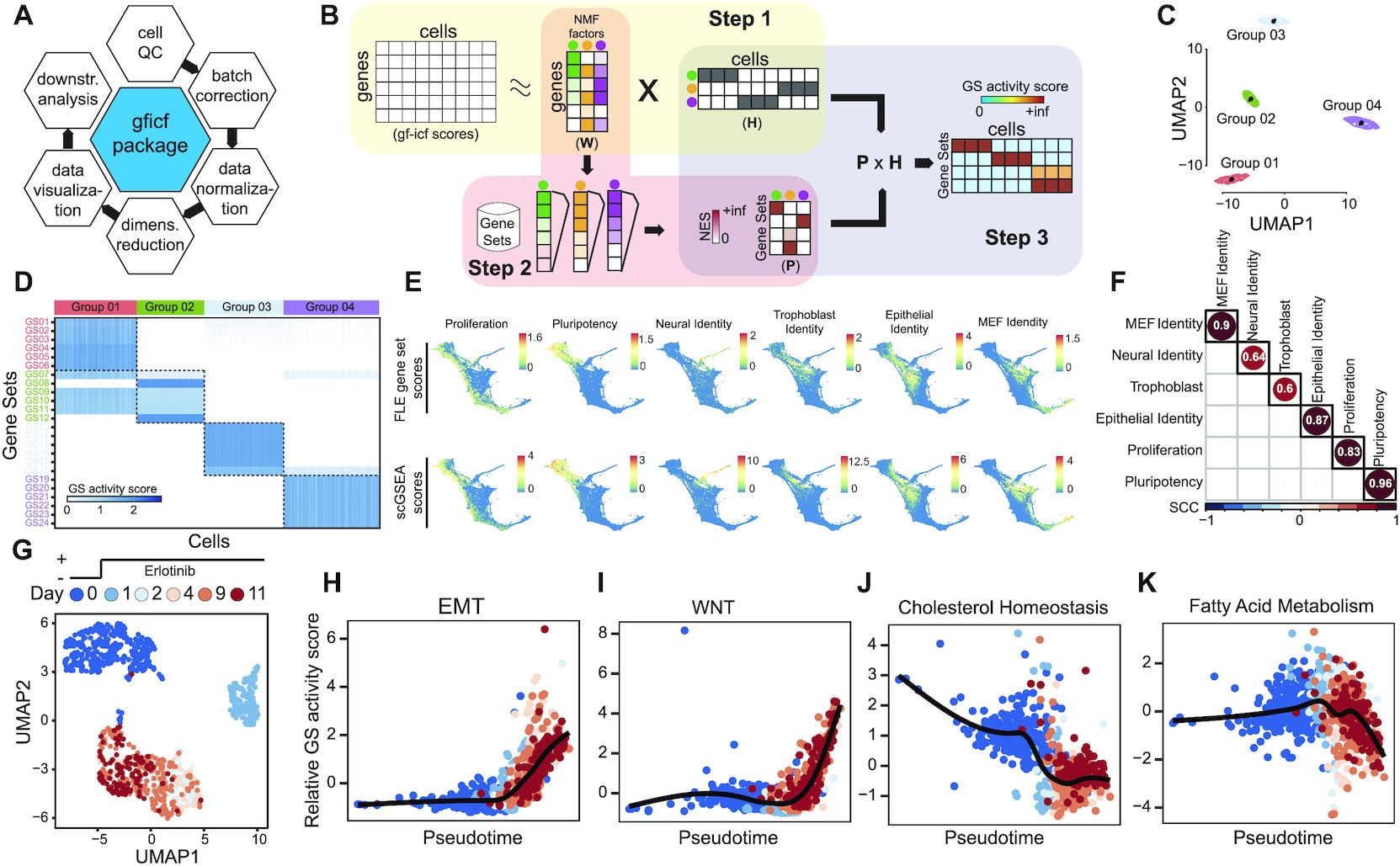
Single-cell gene set enrichment analysis overview and performances. (A) GFICF package overview. (B) Single-cell gene set enrichment analysis pipeline. (C) UMAP plot of 5000 simulated cells grouped in four distinct groups. (D) Reconstructed activity of 24 simulated pathways across the 5000 cells in (C). In the heatmap pathways are along rows while simulated cells along columns. Cells are ordered according to their group of origin. (E) Comparison between scGSEA pathway scores and signature scores originally computed by Schiebinger et al. on 25 1203 single-cell profiles collected during differentiation stages. First row shows original gene set scores computed by Schiebinger et al. using wot phyton package. Second row shows gene set scores computed with scGSEA tool in the gficf R package. Each column represents a different gene set. Scores were plotted on the original FLE (force-directed layout) coordinates published by Schiebinger et al. (F) Spearman Correlation Coefficient (SCC) between scGSEA scores and wot package signature scores across the 25 1203 single-cell transcriptional profiles in (E). (G) UMAP representation of 1044 cells subject to eleven days of consecutive erlotinib treatment. Cells are color-coded according to sequenced day (i.e. 0, 1, 2, 4, 9 and 11 days). Single-cell transcriptional profiles were normalized with gficf package. (H) EMT activity scores against inferred cell pseudo-time using the activity scores of 50 hallmark gene sets downloaded from MSigDB. Cells are color-coded as in (G). (I–K) Same as (H) but for wnt, cholesterol and fatty acid pathways respectively.